Module 3 Data manipulation with R
In Module 2 we introduced the basics of the R language and program environment. In this module, we’ll go a few steps further and get into the finer details of data manipulation: the practice of arranging, sorting, cleaning, and modifying data (and the objects that contain data). Data manipulation is an important R skill because real datasets usually need some fixing before they can be properly analyzed. Data manipulation is also and essential part of R programming for tasks other than data analysis. For example, simulation modeling or document production (e.g., this website!).
3.1 Data import and export
Before you can use R to analyze your data, you need to get you data into R. Most of us store our data in an Excel spreadsheet or some kind of database. These are fine, but if you plan to have R in your workflow I strongly recommend you get in the habit of storing your data in plain text files. Plain text files store data separated (or “delimited”) by a particular character such as a comma or a tab. Unsurprisingly, these are called comma-delimited (.csv) or tab-delimited (.txt) text files. The reason for storing and importing your data this way is simple: plain text files are a stable, almost-universally supported file format that can be read and opened by many programs on any computer. Proprietary formats such as Excel spreadsheets (.xlsx or .xls) or the Mac “Numbers” program (.numbers) are not as widely supported and change more frequently than plain text formats. The fact that plain text formats are typically smaller than equivalent Excel files is a nice bonus.
Reading your data in from text files has another benefit: repeatability. Chances are you will need to revisit or repeat your data analysis several times. If you program the importation of data in a few lines of R code, you can just run the code. If, however, you use one of the “point-and-click” methods available in RStudio or base R you will have to repeat those steps manually. This can get very aggravating and time consuming if you have multiple datasets to import. This page is designed to demystify the process of getting data into R and to demonstrate some good practices for data storage.
Note that many of the code blocks below have # not run: in them, and no output. These indicate that either single commands or the entire block was not actually evaluated when rendering the R Markdown. This was either because the code was designed to return an error (which would prevent R Markdown rendering), produce large outputs, or would clutter up the folder where I store the files for this website. These commands will work on your machine, and will demonstrate important R concepts, so try them out!
3.1.1 Importing data: preliminaries
Before you try to import your data into R, take a few minutes to set yourself up for success.
3.1.1.1 Naming data files
Good file names are essential for a working biologist. Chances are you will work on many projects over the course of your career, each with its own collection of data files. Even if you store each project’s data in a separate folder, it is a very good idea to give data files within those folders names that are descriptive. I have worked with many people who insist on using file names like “data.xlsx” and relying on their own memory or paper notes to tell them which file is which. Consider this list of particularly egregious file names:
- data.txt
- Data.txt
- data.set.csv
- analysis data.xlsx
- thesis data.xlsx
- DATA FINAL - copy.txt
- data current.revised.2019.txt.xlsx
- final thesis data revised revised final revised analysis.csv
We are all guilty of this kind of bad naming from time to time. What do all of the above have in common? They are vague. The names contain no meaningful information about what the files actually contain. Some of them are ambiguous: different operating systems or programs might read “data.txt” and “Data.txt” as the same file, or as different files!
Data file names should be short, simple, and descriptive. There are no hard and fast rules for naming data files so I’m not going to prescribe any. Instead, I’ll share some examples and explain why these names are helpful. As you go along and get getter at working with data you will probably develop your own habits, and that’s ok. Just try to follow the guiding principles of short, simple, and descriptive.
Here are some example file names with explanations of their desirable properties:
sturgeon_growth_data_2017-02-21.txt
This name declares what the file contains (growth data for sturgeon), and the date it was created. Notice that the date is given as “YYYY-MM-DD”. This format is useful because it automatically sorts chronologically. This file name separates its words with underscores rather than spaces or periods. This can prevent some occasional issues with programs misreading file names (white space and periods can have specific meanings in some contexts).
dat_combine_2020-07-13.csv
This name signifies that the file contains a “combined” dataset, and includes the date of its creation. This would be kept in a folder with the files that were combined to make it. Within a project I usually use prefixes on file names to help identify them: “dat” for datasets with response and explanatory variables; “met” for “metadata”; “geo” or “loc” for geospatial datasets and GIS layers; and “key” for lookup tables that relate dat and met files. You might develop your own conventions for types of data that occur in your work.
dat_vegcover_nrda_releaseversion_2021-05-03.csv
This name is similar to the last name, but includes some additional signifiers for the project funder (NRDA) and the fact that this is the final released version.
3.1.1.2 Formatting data files
Recently Broman and Woo (2018) reviewed common good practices for storing data in spreadsheets. Their article is short and easy to read, and gave a few recommendations for data formatting:
- Be consistent
- Choose good names for things
- Write dates as YYYY-MM-DD
- No empty cells
- Put just one thing in a cell
- Make it a rectangle
- Create a data dictionary
- No calculations in the raw data files
- Do not use font color or highlighting as data
- Make backups
- Use data validation to avoid errors
- Save the data in plain text files
Most people who work with data, myself included, probably agree with their recommendations. There is room for individual preferences in the exact implementation of each principle, but the general ideas are solid. You will save yourself a lot of headache if you follow their advice. I strongly suggest you read their paper and follow their advice.
3.1.1.2.1 Variable names
Variable names, like data file names, should be brief but informative. There are few restrictions on what variable names can be in R. For simplicity, I recommend you use only lowercase letters, numbers, underscores (_), and periods (.) in variable names. Other symbols are either inconvenient to type, or can cause issues being parsed by R. Some common pifalls include:
Leading numerals will cause an error if not enclosed in quotation marks:
Numeral-only names will cause an error. They can also be misinterpreted as column numbers. Don’t give your variables numeral-only names.
x <- iris[1:5,]
names(x)[1] <- "3"
x$3 # error
x$'3' # works
x[,3] # column number 3
x[,"3"] # column named "3"Spaces in variable names will cause errors because R interprets a space as whitespace, not part of a name. If you insist on using spaces, enclose the variable name in quotation marks every time you use it. This is one of the few places where R cares about whitespace.
## [1] 5.1 4.9 4.7 4.6 5.0Special characters in variable names can cause issues with parsing. Many symbols that you might think to use, such as hyphens (aka: dashes, -) or slashes / are also R operators. Again, try to avoid these issues by using only letters, numbers, underscores, and periods.
## [1] 5.1 4.9 4.7 4.6 5.0## surv% Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa## [1] 5.1 4.9 4.7 4.6 5.0Uppercase letters in variable names work just fine, but they can be inconvenient. Because R is case sensitive, you need to type the variable exactly the same way each time. This can be tedious if you have several variables with similar names. In my code I prefer to use only lower case letters, so I don’t ever have to remember how names are capitalized.
x <- iris[1:5,]
names(x)[1] <- "sep.len" # convenient
names(x)[2] <- "SEP.LEN" # slightly inconvenient
names(x)[3] <- "Sep.Len" # more inconvenient
names(x)[4] <- "SeP.lEn" # just don't
x## sep.len SEP.LEN Sep.Len SeP.lEn Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa3.1.1.2.2 Data values
All data in R have a type: numeric, logical, character, etc. Within a vector, matrix, or array, all values must have the same type or they will be converted to character strings. Each variable in a data frame (the most common R data structure) is essentially a vector. This means that in your input file you should try to have one and only one type of data in each column. Otherwise, R will coerce the values into character strings, which may or may not be a problem. Putting only one type of value into each column will also help get your data into “wide” or “tidy” format (Wickham (2014)).
Entering data should also include some measure of quality assurance and quality control (QA/QC). This basically means making sure that your data are entered correctly, and are formatted correctly. Correct entry can be accomplished by direct comparison to original datasheets or lab notebooks. Cross-checking is a valuable tool here: have two people enter the data and compare their files.
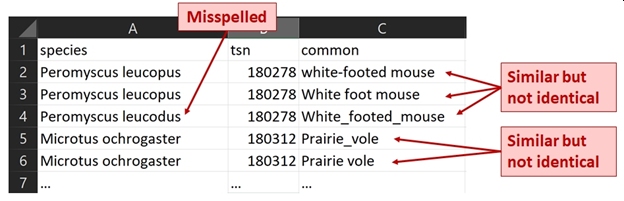
Data formatting requires thinking about what kinds of data are being entered. For numbers, this means entering the correct values and an appropriate number of significant digits. For text values, this may mean spell-checking or settling on a standard set of possible values. The figure below shows some problems that a QA/QC step might fix:
3.1.2 Importing data from text files with read.csv() and read.table()
Plain text files are imported, or read, using functions that are named for that purpose. The function read.table() works on many types of text files, including tab-delimited and comma-delimited. The function read.csv() works on comma-delimited files only. The syntax for both is very similar.
Put the files elk_data_2021-01-21.txt and elk_data_2021-01-21.csv in your R home directory. The R home directory depends on your operating system:
- Windows users: the home directory is your Documents folder. The address of this folder is
C:/Users/username/Documents, whereusernameis your Windows logon name. You can get to your Documents folder by opening Windows Explorer (Win-E) and double-clicking “Documents”. - Mac users: the home directory is your home folder. The address of this folder is
/Users/username(whereusernameis your username) or~/. You can open your home folder by pressing CMD-SHIFT-H, or by using the Go pulldown menu from the menu bar.
Import a tab-delimited file from your home directory:
Import a comma-delimited version from your home directory. Note how similar the syntax is to the first command.
dat <- read.csv("elk_data_2021-01-21.csv", header=TRUE)
# alternative, using `read.table()` with
# sep argument to specify how values are separated:
dat <- read.table("elk_data_2021-01-21.csv",
header=TRUE, sep=",")The examples above required the name of the data file—-notice the file extensions!—-and an argument header which specifies that the first row of the data file contains column names. Try one of those commands without that argument and see what happens.
## height antler species
## 1 69.3 24.8 unknown
## 2 69.3 34.5 unknown
## 3 87.7 53.5 unknown
## 4 99.8 58.3 unknown
## 5 69.3 61.2 unknown
## 6 84.1 67.3 unknown## V1 V2 V3
## 1 height antler species
## 2 69.3 24.8 unknown
## 3 69.3 34.5 unknown
## 4 87.7 53.5 unknown
## 5 99.8 58.3 unknown
## 6 69.3 61.2 unknownWithout the header argument, the column names were read as the first row of the dataset and the columns of the data frame were given generic names V1, V2, and so on. As a side effect, the values in each of the columns are all of character type instead of numbers. This is because both read.table() and read.csv() import all values as character strings by default, and convert to other types only if possible. In this case, conversion is not possible because the strings height, antler, and species cannot be encoded as numbers, but the numbers form the original file can be encoded as character strings. Further, the values within a single column of a data frame are a vector, and all elements of a vector must have the same type. Thus, importing without the header argument will likely result in a data frame full of text representations of your data.
Both read.table() and read.csv() will bring in data stored as a table, and produce a type of R object called a data frame. This data frame must be saved to an object to be used by R. If not saved, the data frame will be printed to the console. Compare the results of the two lines below on your machine.
read.table("elk data 2021-01-21.txt", header=TRUE)
dat <- read.table("elk data 2021-01-21.txt", header=TRUE)The first command read the file, and printed it to the console. The second command read the file and stored it in an object named dat. Nothing is returned to the console when this happens.
3.1.2.1 Important arguments to read.table() and read.csv()
3.1.2.1.1 file
The first argument to read.table() and read.csv() is file, which is the filename, including directory path, of the file to be imported. By default, R will look in your home directory for the file. If the file is stored in the home directory, then you can just supply the filename as a character string. Don’t forget the file extension! Most of the examples on this site will, for convenience, use data files stored in your home directory. In your own work you may want to use other folders.
If you want to import a file from another folder, you need to supply the folder name as part of the argument file. My preferred way to do this is by specifying the folder and file name separately, and combining them with the paste() command. The function paste() concatenates, or sticks together, character strings. Separating the folder from the file name also makes your code more portable. For example, if you needed to run your code on another machine you would only need to update the directory. An example of how this might work is shown below:
# not run:
# directory that contains all data and folders
top.dir <- "C:/_data"
# project folder
proj.dir <- paste(top.dir, "ne stream/dat", sep="/")
# folder that holds dataset
data.dir <- proj.dir
# name of dataset
data.name <- "dat_combine_2020-07-13.csv"
# import dataset
in.name <- paste(data.dir, data.name, sep="/")
dat <- read.csv(in.name, header=TRUE)In the example above, moving to a new machine would only necessitate changing one line, the definition of data.dir (or top.dir, if you keep the same folder structure on multiple machines).
3.1.2.1.2 header
We’ve already seen the importance of header. You’ll use header=TRUE in almost every import command you ever run. Occasionally you’ll run into datasets that do not come with a header row. Of course, you won’t store your data that way because it complicates data management.
3.1.2.1.3 sep
As seen in one of the examples above, the sep argument specifies what character separates values on each line of the data file. This argument has some useful default values. If you are using read.table() with a tab-delimited file, or read.csv() with a comma-delimited file, you don’t need to set sep.
The default in read.table() reads any white space as a delimiter. This is fine if you have a tab-delimited file, but if the file contains a mix of spaces and tabs you might have some problems. For example, if entries are separated by tabs, but some values contain spaces. In those cases, specify the appropriate delimiter:
# not run:
in.name <- "elk_data_2021-01-21.txt"
# tab-delimited (sep="\t"):
dat <- read.table(in.name, header=TRUE, sep="\t")
# space-delimited (sep=" "):
#dat <- read.table(in.name, header=TRUE, sep=" ")The sep argument is usually necessary when using the “clipboard trick” (see below).
3.1.2.1.4 row.names
When R imports a table of data, it will assign names to the rows. The default names are just the row numbers. Some data files will have their own row names. If you want to not import these names, you can set row.names argument to FALSE to get the default numbers. You can also specify names as a character vector.
3.1.2.1.5 col.names
The argument col.names works in a similar manner to row.names, but for column names. If column names are not provided with argument header, R will assign the columns V followed by a column number (V1, V2, etc.). If you want to provide your own names, use col.names. Personally, I’ve never used this argument and instead prefer to either include column names in my data files or just assign names after importing.
3.1.2.1.6 skip
Some data files will contain rows of text or other information before the tabular data starts. The argument skip allows you to skip these lines. Without skip, R will try to read those rows of text as if they were data and fail to import. Use the file elk_data_2021-07-14.txt, which has some extra rows of text that would otherwise cause read.table() to fail.
# not run:
# alternate file with some header rows
in.name <- "elk_data_2021-07-14.txt"
# fails:
dat <- read.table(in.name, header=TRUE)
# works:
dat <- read.table(in.name, header=TRUE, skip=2)
datThe error message when you tried to import the file without skip=2 comes up a lot when trying to import data into R. What is happening is that R uses the number of elements in the first row to calculate the dimensions of the data frame that it makes upon import. In this case, it interpreted the 7 words in the first row of text, Here is some text about the dataset, as 7 column headings Here, is, some, text, about, the, and dataset. Then, when next line that came from Gould (1974). only had 5 elements (that, came, from, Gould, and (1974)), this caused an error.
Why does this issue vex so many new R users (and experienced R users)? Often we will have incomplete records in a data set, where a row is missing values in one or more columns. Sometimes blank values are not read correctly. Consider the file elk_data_blanks_2021-07-14.txt, which has a blank in line 13.
When we try to import that file, we get a similar error:
There are a few ways to deal with this error. The first is to go back to the original datafile and put R’s blank value, NA, in the blanks. You could also put in a value that is not likely to occur, such as -9999 in variables that must be positive. This would make your file more readable, but without locking you in to using R. Either solution will ensure that missing values are not interpreted as extra white space. This method can be very time consuming for very large datasets, even in Excel (or especially in Excel). The best time to fix this issue is at data entry. Recall that having no empty cells is one of the recommendations of Broman and Woo (2018).
Another way is to save the file as a comma-delimited file, because commas are less ambiguous than white space for defining missing values. This method usually works, but might still create problems if there are multiple blank values per record or blanks at the ends of rows. The third option is to use the fill argument in the R import functions (see below).
3.1.2.1.7 fill
As described above, missing values can cause problems when importing data into R. One way to deal with missing values that occur at the end of a line in the data file is to use the fill argument. Setting fill=TRUE will automatically pad the data frame with blanks so that every row has the same number of values.
As convenient as fill=TRUE can be, you need to be careful when using it. R may add the wrong number of values, or values of the wrong type. In the example above, the value of species in row 13 is a blank character string (""), not a missing value NA. This is an important difference because "" and NA are treated very differently by many R functions. The commands below show that R interprets the padded entry "" as a character string of length 0, not a missing value.
##
## alces megaloceros unknown
## 1 2 1 16## Group.1 x
## 1 107.8000
## 2 alces 164.4500
## 3 megaloceros 239.1000
## 4 unknown 85.1375## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE3.1.2.1.8 stringsAsFactors
A factor is a variable in a dataset that defines group membership: control vs. treated, red vs. blue, gene 1 vs. gene 2 vs. gene 3, and so on. The different groups to which an observation can belong are called the levels of the factor. This is very useful for statistical modeling, but can be a pain in the neck when trying to work with data in R. Functions read.table() and read.csv() have an argument to automatically convert character strings to factors for convenience: stringsAsFactors. This is a logical switch that will interpret character strings as levels of a factor (stringsAsFactors=TRUE) or as character strings (stringsAsFactors=FALSE). The default was TRUE until around version 4.0 or so, but now the default is the much more convenient FALSE.
The reason that factors can cause headaches in R is because although they look like character strings, they are really numeric codes and the text you see is just a label. If you try to use a factor as if it was a character string, you can get strange errors. My advice is to always import data with stringsAsFactors=FALSE and only convert to factor later if needed. Most statistical functions will convert to factor for you automatically anyway. The only time when you may want to store data as a factor is when you a very large dataset with many levels of a factor; in those situations, factors will use memory more efficiently.
If you get tired of typing stringsAsFactors=TRUE in your import commands, you can set an option at the beginning of your script that will change the default behavior of read.table() and read.csv():
Back when this option was TRUE by default I put this command at the start of every new R script to avoid problems with text strings being misinterpreted as factors.
3.1.3 Importing data from saved workspaces
3.1.3.1 Importing R objects with readRDS()
Some R objects are not easily represented by tables. For example, the outputs of statistical methods are usually lists. Such objects can be saved using function saveRDS() and read into R using readRDS(). The example below will save an R data object to file rds_test.rds in your R working directory.
##
## Call:
## lm(formula = Petal.Width ~ Sepal.Width, data = iris)
##
## Coefficients:
## (Intercept) Sepal.Width
## 3.1569 -0.6403##
## Call:
## lm(formula = Petal.Width ~ Sepal.Width, data = iris)
##
## Coefficients:
## (Intercept) Sepal.Width
## 3.1569 -0.6403There is a lazier way to accomplish this task using function dput(). This function saves a plain text representation of an object that can later be used to recreate the object (i.e., R code that can reproduce the object). The help file for dput() cautions that this is not a good method for transferring objects between R sessions and recommends using saveRDS() instead. That being said, I’ve used this method for years and have never had a problem. The procedure is to copy the output of dput() into your R script, and later assign that text to another object. Use dput() at your own risk.
## structure(list(Sepal.Length = c(5.1, 4.9, 4.7, 4.6, 5, 5.4),
## Sepal.Width = c(3.5, 3, 3.2, 3.1, 3.6, 3.9), Petal.Length = c(1.4,
## 1.4, 1.3, 1.5, 1.4, 1.7), Petal.Width = c(0.2, 0.2, 0.2,
## 0.2, 0.2, 0.4), Species = structure(c(1L, 1L, 1L, 1L, 1L,
## 1L), levels = c("setosa", "versicolor", "virginica"), class = "factor")), row.names = c(NA,
## 6L), class = "data.frame")## structure(list(Sepal.Length = c(5.1, 4.9, 4.7, 4.6, 5, 5.4),
## Sepal.Width = c(3.5, 3, 3.2, 3.1, 3.6, 3.9), Petal.Length = c(1.4,
## 1.4, 1.3, 1.5, 1.4, 1.7), Petal.Width = c(0.2, 0.2, 0.2,
## 0.2, 0.2, 0.4), Species = structure(c(1L, 1L, 1L, 1L, 1L,
## 1L), levels = c("setosa", "versicolor", "virginica"), class = "factor")), row.names = c(NA,
## 6L), class = "data.frame")## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa3.1.3.2 Importing R workspaces with load()
Everything you do in R takes place within the workspace. This can be thought of as a sandbox where all of your data and functions reside. If you want to save this entire environment and continue working later you can do this by saving the workspace (see below). Workspaces can be loaded using function load(). The example below loads workspace example.RData from your home directory. Notice that this command will load every object from the saved workspace into your current workspace. This might cause problems if the saved workspace and current workspace have objects with the same names.
When loading a workspace, it is sometimes more useful to import it into a new environment. This will let you import objects from that workspace without putting its objects into your current workspace. I use this method a lot for working with model outputs. The typical workflow is:
- Run an analysis or set of simulations in multiple R instances and save the results of each in its own R workspace.
- After simulations are complete, import the results of each simulation from its individual workspace into a new environment.
- Copy the objects from the environment containing the loaded workspace to the main workspace.
- Delete the environment containing the loaded workspace.
- Repeat until all results are imported.
Here is an example of importing individual objects from a workspace into a new environment, then copying that object into the current workspace:
3.1.4 Importing data: special cases:
3.1.4.1 Getting data from the clipboard
Sometimes it can be convenient to copy and paste data directly from a spreadsheet into R. This is fine for quick or one-off data imports, but not recommended for routine use because it is tedious to reproduce and error-prone. The procedure is as follows:
- In the R console, type this command but do NOT run it:
a <- read.table("clipboard", header=TRUE, sep="\t") - In Excel, highlight your data and copy it to the clipboard using Ctrl-C.
- Click in the R console to move your focus there. In R, hit the ENTER key.
The command you typed in step 1 will read the table from the clipboard. Note that if you try to copy/paste the read.table("clipboard",...) command into the R console, you will then have that command in your clipboard and not your data.
3.1.4.2 Entering data using scan()
The scan() function lets you enter values one-by-one, separated by pressing ENTER. You can use this just like typing values into Excel. Or, you can copy and paste a single column of values from Excel. The latter case works because R interprets the values within a column as being separated by a line return (i.e., pressing ENTER).
Type the name of an object to which data will be saved, the assignment operator <-, scan(), then ENTER, then values (each followed by ENTER), then hit ENTER twice after the last value. This is obviously inefficient for large datasets, but it’s a nice trick to be aware of.
To enter values from a column in Excel, highlight and copy the column. Then go to R, enter the command below, and press ENTER. Then paste your values and press ENTER again.
3.1.4.3 Entering data directly using c()
Function c() combines its arguments as a vector. As with all functions, arguments must be separated by commas. Arguments can be on the same line or on different lines.
## [1] 1 2 3 9 8 7 6 5## [1] 1 3 5Like scan(), data can be entered directly into the console using c(). Also like scan(), this is comically inefficient but can be useful for small numbers of values.
3.1.5 Export data from R
3.1.5.1 Writing out tables (best method)
Everything you do in R takes place within the “workspace”. You can save entire workspaces, but in most situations it is more useful to write out data or results as delimited text files. “Delimited” means that individual data within the file are separated, or delimited, by a particular symbol. The most common formats are comma-delimited files (.csv) or tab-delimited files (.txt). Which format to use is largely a matter of personal preference. I’ve found that comma-delimited files are slightly less likely to have issues because they use a character (,) rather than whitespace to separate values.
Tab-delimited files are written out using write.table(). In the example below, the first argument dat is the object from the R workspace that you want to write out. The argument sep works the same way as in read.table(). The example below will save a file called mydata.txt in your home directory (R’s working directory). Option sep="\t" can be provided to ensure that the file is written as a tab-delimited file and not a space-delimited file.
Comma-delimited files are written out by write.csv(). The syntax is similar to write.table(). Note that the file extension must be “.csv” instead of “.txt”. If you save a file with the wrong extension, then it may be corrupt or unusable (or, R might return an error).
Most of the time you will not be saving files to your home directory, but to another folder. The folder and the file name must both be specified to save somewhere else. It is usually a good idea to define the file name and file destination separately. This way if you re-use your code on another machine, or a different folder, you only need to update the folder address once.
# not run:
# do this:
out.dir <- "C:/_data"
out.name <- "mydata.csv"
write.csv(dat, paste(out.dir, out.name, sep="/"))
# or this:
out.dir <- "C:/_data"
out.name <- "mydata.csv"
out.file <- paste(out.dir, out.name, sep="/")
write.csv(dat, out.file)
# not this:
write.csv(dat, "C:/_data/mydata.csv")The latter example is problematic because the output file name contains both the destination folder and the filename itself. If you try to run your code on another machine, it will take longer to update the code with a new folder address. This method is also problematic because exporting multiple files will require typing the folder address multiple times. Consider this example:
# not run:
dat1 <- iris[1:6,]
dat2 <- iris[7:12,]
write.csv(dat1, "C:/_data/mydata1.csv")
write.csv(dat2, "C:/_data/mydata2.csv")Running that code on a new machine, or modifying it for a new project in a different folder, requires changing the folder address twice. The better way is to define the folder name only once:
# not run:
out.dir <- "C:/_data"
write.csv(dat1, paste(out.dir, "mydata1.csv", sep="/"))
write.csv(dat2, paste(out.dir, "mydata2.csv", sep="/"))See the difference? The revised code is much more portable and reusable. This illustrates a good practice for programming in general (not just R) called “Single Point of Definition”. Defining variables or objects once and only once makes your code more robust to changes and easier to maintain or reuse. The example below shows a more compact way to export the files mydata1.csv and mydata2.csv, with even fewer points of definition for file names:
# not run:
out.dir <- "C:/_data"
out.names <- paste0("mydata", 1:2, ".csv")
out.files <- paste(out.dir, out.names, sep="/")
write.csv(dat1, out.files[1])
write.csv(dat2, out.files[2])One handy trick is to auto-generate an output filename that includes the current date. This can really help keep multiple versions of analyses or model runs organized. The paste0() command pastes its arguments together as they are, with no separator between them (equivalent to paste(...,sep="")). Function Sys.Date() prints the current date (according to your machine) in YYYY-MM-DD format.
3.1.5.2 Writing out individul R objects
Some data do not fit easily into tables. Individual R objects can be saved to an “.rds” file using the function saveRDS(). This function saves a single object that can later be loaded into another R session with function readRDS().
3.1.5.3 Saving entire R workspaces
Entire R workspaces or parts of workspaces can be saved using the function save.image(). The command by itself will save the entire current workspace with a generic name to the working directory. It is more useful to save the workspace under a particular name and/or only save specific objects.
3.1.5.4 Writing text to a file (special cases)
Sometimes you need to write out data that do not fit easily into tables. An example might be the results of a single t-test or ANOVA. This is also sometimes necessary when working with programs outside of R. For example, the programs OpenBUGS and JAGS require a text file that defines the model to be fit. This text file can be generated by R, which makes your workflow simpler because you don’t need to use a separate program to store the outputs.
The function to write such files is sink(). Functions print() and cat() are also useful in conjunction with sink().
sink()works in pairs. The first command defines a file to put text into. The second command finishes the file and writes it.print()prints its arguments as-is. It should produce the same text as running its argument in the R console.cat()stands for “concatenate and print”. Use this to combine expressions and then print them.
3.2 Making values in R
The typical analytical workflow in R involves importing data from text files or other sources. Sometimes, however, it is necessary to make values within R. This can be for a variety of reasons:
- Producing test data sets with desired distributions
- Making sets of indices or look-up values to use in other functions
- Modifying existing datasets
- Running simulations
- And many more…
This section demonstrates some methods for producing values–i.e., data–within R.
3.2.1 Producing arbitrary values with c()
In a previous section we saw how function c() combines its arguments as a vector. As with all functions, arguments must be separated by commas and can be on different lines. This method can be used to make vectors with any set of values.
## [1] 1 2 3 9 8 7 6 5Like scan(), this is an inefficient way to make vectors but can be useful for small numbers of values. Or, for sets of values that can’t be defined algorithmically.
3.2.2 Generating regular values
3.2.2.1 Consecutive values with :
Consecutive values can be generated using the colon operator :. A command like X:Y will generate consecutive values from X to Y in increments of 1 or -1. If \(X<Y\), the increment is 1; if \(X>Y\), the increment is -1. The usual use case is to make sets of consecutive integers.
## [1] 1 2 3 4 5 6 7 8 9 10## [1] 5 4 3 2## [1] -1 0 1 2 3 4 5 6 7 8 9 10## [1] 10 9 8 7 6 5 4 3 2 1 0 -1Sometimes you may need to use parentheses to get the intended values because of the order in which R runs operators (aka: “operator precedence”).
## [1] 5## [1] 5 10## [1] 1 2 3 4 5 6 7 8 9 10X and Y don’t have to literally be numbers; they can be variables or R expressions that evaluate to a number. Below are some examples:
## [1] 5 6 7 8 9 10Most people use : to produce sets of integers, but it can also make non-integers. The values will be separated by 1 or -1. The first value will be the value before the :. The last value might not be the value after the :. It will be the value up to or before the second number along the sequence from the first number. If the value before the : is an integer, then the sequence will consist of integers. Note that if you want a sequence that includes non-integers, it might be safer to use seq() (see below).
## [1] 2.3 3.3 4.3## [1] 5 4 3## [1] 2.7 3.7 4.7 5.73.2.2.2 Regular sequences with seq()
Function seq() generates regular sequences of a given length (length=), or by an interval (by=). The command must be supplied with a length or an interval, but not both. If you specify a length, R will calculate the intervals for you. If you specify an interval, R will calculate the length for you. If you specify a length and an interval, R will return an error.
## [1] 0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5 6.0 6.5 7.0
## [16] 7.5 8.0 8.5 9.0 9.5 10.0 10.5 11.0 11.5 12.0 12.5 13.0 13.5 14.0 14.5
## [31] 15.0 15.5 16.0 16.5 17.0 17.5 18.0 18.5 19.0 19.5 20.0## [1] -3.14159265 -3.04159265 -2.94159265 -2.84159265 -2.74159265 -2.64159265
## [7] -2.54159265 -2.44159265 -2.34159265 -2.24159265 -2.14159265 -2.04159265
## [13] -1.94159265 -1.84159265 -1.74159265 -1.64159265 -1.54159265 -1.44159265
## [19] -1.34159265 -1.24159265 -1.14159265 -1.04159265 -0.94159265 -0.84159265
## [25] -0.74159265 -0.64159265 -0.54159265 -0.44159265 -0.34159265 -0.24159265
## [31] -0.14159265 -0.04159265 0.05840735 0.15840735 0.25840735 0.35840735
## [37] 0.45840735 0.55840735 0.65840735 0.75840735 0.85840735 0.95840735
## [43] 1.05840735 1.15840735 1.25840735 1.35840735 1.45840735 1.55840735
## [49] 1.65840735 1.75840735 1.85840735 1.95840735 2.05840735 2.15840735
## [55] 2.25840735 2.35840735 2.45840735 2.55840735 2.65840735 2.75840735
## [61] 2.85840735 2.95840735 3.05840735## [1] -1.00000000 -0.89473684 -0.78947368 -0.68421053 -0.57894737 -0.47368421
## [7] -0.36842105 -0.26315789 -0.15789474 -0.05263158 0.05263158 0.15789474
## [13] 0.26315789 0.36842105 0.47368421 0.57894737 0.68421053 0.78947368
## [19] 0.89473684 1.00000000If you want the values to go from greater to smaller, the by argument must be negative.
## [1] -1 -2 -3 -4 -5 -6 -7 -8 -9 -103.2.2.3 Repeated values with rep()
The function rep() creates a sequence of repeated elements. It can be used in several ways.
One way is to specify the number of times its first argument gets repeated. Notice that in the second command rep(1:3, 5), the entire first argument 1 2 3 is repeated 5 times.
## [1] 2 2 2 2 2## [1] 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3Another way is to repeat each element of its input a certain number of times each:
## [1] 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3The arguments times and each can be combined. Notice that each takes precedence over times.
## [1] 1 1 1 1 2 2 2 2 3 3 3 3 1 1 1 1 2 2 2 2 3 3 3 3## [1] 1 1 2 2 3 3 1 1 2 2 3 3 1 1 2 2 3 3 1 1 2 2 3 33.2.2.4 The “Recycling Rule”
One quirk of R sequences and vectors is the recycling rule. Values in one vector can be “recycled” to match up with values in another vector. This rule is only invoked if the length of the longer vector is a multiple of the length of the shorter vector. Otherwise you will get an error. Consider the following example:
my.df <- data.frame(x=1:12)
# add a value with 3 unique values
my.df$x3 <- 1:3
# add a value with 4 unique values
my.df$x4 <- 1:4
# add a value with 2 random values:
my.df$x2 <- runif(2)
my.df## x x3 x4 x2
## 1 1 1 1 0.9373858
## 2 2 2 2 0.8504828
## 3 3 3 3 0.9373858
## 4 4 1 4 0.8504828
## 5 5 2 1 0.9373858
## 6 6 3 2 0.8504828
## 7 7 1 3 0.9373858
## 8 8 2 4 0.8504828
## 9 9 3 1 0.9373858
## 10 10 1 2 0.8504828
## 11 11 2 3 0.9373858
## 12 12 3 4 0.8504828Trying to add a variable with a number of values that is not a factor of 12–5, 7, 8, 9, 10, or 11–will usually return an error.
3.2.3 Generating random values
R can generate many types of random values. This makes sense because R was designed for statistical analysis, and randomness is a key feature of statistics. When pulling random values in R (or any other program, for that matter) it is advisable to set the random number seed so that your results are reproducible. The random number seed is needed because “random” numbers from a computer are not actually random. They are pseudo-random, which means that while the values appear random, and can pass many statistical tests for randomness, they are calculated deterministically from the state of your computer. If you are running simulations, or an analysis that involves random sampling, you need to set the random number seed. Otherwise, you might get different results every time you run your code. This can make reproducing an analysis or set of simulations all but impossible.
The random number seed can be set with function set.seed(). Setting the seed defines the initial state of R’s pseudo-random number generator.
# different results each time:
runif(3)
## [1] 0.5798209 0.8214039 0.1137186
runif(3)
## [1] 0.7645078 0.6236135 0.1484466
runif(3)
## [1] 0.08026447 0.46406955 0.77936816
# same results each time:
set.seed(42)
runif(3)
## [1] 0.9148060 0.9370754 0.2861395
set.seed(42)
runif(3)
## [1] 0.9148060 0.9370754 0.2861395
set.seed(42)
runif(3)
## [1] 0.9148060 0.9370754 0.28613953.2.3.1 Random values from a set with sample()
The function sample() pulls random values from a set of values. This can be done with or without replacement. If you try to pull too many values from a set without replacement R will return an error. The default argument for replace= is FALSE.
set.seed(456)
sample(1:10, 5, replace=FALSE)
## [1] 5 3 6 10 4
sample(1:10, 5, replace=TRUE)
## [1] 9 10 9 10 3You can use sample() with values other than numbers. The function will pull values from whatever its first argument is.
x1 <- c("heads", "tails")
sample(x1, 10, replace=TRUE)
## [1] "tails" "heads" "tails" "tails" "tails" "heads" "tails" "heads" "heads"
## [10] "tails"
x2 <- c(2.3, pi, -3)
sample(x2, 10, replace=TRUE)
## [1] 2.300000 2.300000 2.300000 3.141593 3.141593 -3.000000 -3.000000
## [8] 3.141593 3.141593 2.300000By default, every element in the set has an equal probability of being chosen. You can alter this with the argument prob. This argument must have a probability for each element of the set from which value are chosen, and those probabilities must add up to 1.
3.2.3.2 Random values from probability distributions
One of the strengths of R is its ease of working with probability distributions. We will explore probability distributions more in Module 5, but for now just understand that a probability distribution is a function that describes how likely different values of a random variable are. In R, random values from a probability distribution can be drawn using a function with a name like r_(), where _ is the (abbreviated) name of the distribution.
The syntax of the random distribution functions is very consistent across distributions. The first argument is always the sample size (n), and the next few arguments are the distributional parameters of the distribution. These arguments can be provided with or without their names. The default order of the arguments is shown on the help page for each function (e.g., ?rnorm). Below are some examples of random draws from different distributions, displayed as histograms.
# sample size
N <- 1000
# draw random distributions
## normal with mean 0 and SD = 1
a1 <- rnorm(N)
## normal with mean 25 and SD = 100
a2 <- rnorm(N, 25, 100)
## uniform in [0, 1]
a3 <- runif(N)
## uniform in [5, 25]
a4 <- runif(N, 5, 25)
## Poisson with lambda = 10
a5 <- rpois(N, 10)
## lognormal with logmu = 1.2 and logsd = 2
a6 <- rlnorm(N, 1.2, 0.9)
# produce histogram of each distribution above
par(mfrow=c(2,3))
hist(a1)
hist(a2)
hist(a3)
hist(a4)
hist(a5)
hist(a6)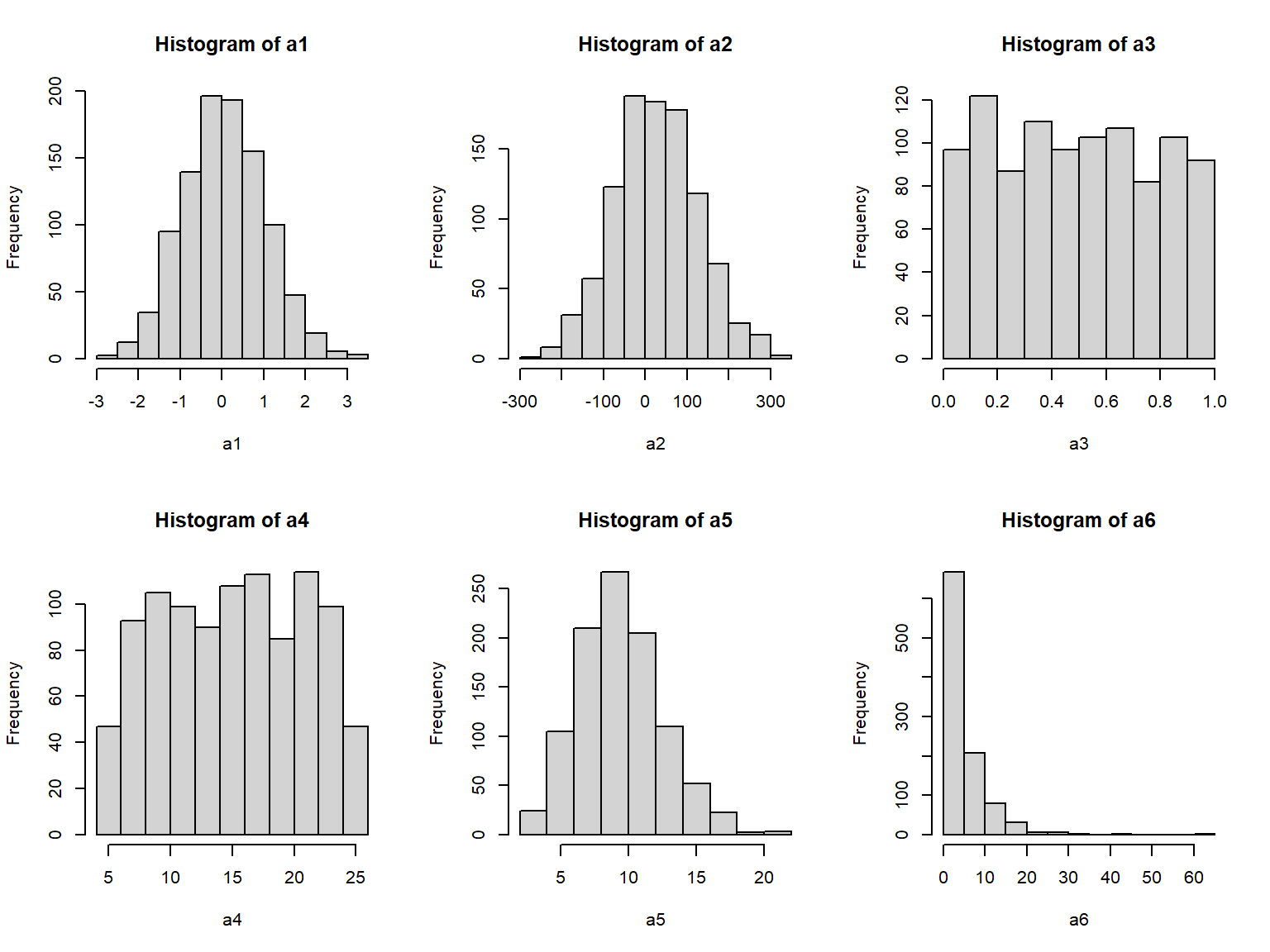
3.3 Selecting data with []
One of the most common data manipulation tasks is selecting, or extracting, from your data. The most common way of selecting subsets of an object (aka: extracting) is the R bracket notation. This refers to the symbols used, []. These are also sometimes called square brackets, to distinguish them from curly brackets, {}. The latter are more properly called braces and have a different purpose in R.
Brackets tell R that you want a part of an object. Which part is determined either by a name, or an index inside the brackets. Indices should usually be supplied as numbers, but other types can be used. For objects with more than 1 dimension, you need to supply as many indices, or vectors of indices, as the object has dimensions. Within brackets, vectors of indices are separated by commas.
Brackets can also be used to exclude certain elements by using the negative sign, but this can be tricky. We’ll explore how negative indices work below.
3.3.1 Basics of brackets
3.3.1.1 Selecting from a vector
Vectors have 1 dimension, so you must specify a single vector of indices. Notice that the brackets can be empty, in which case all elements can be returned. If the vector supplied is of length 0, then no elements will be returned. When a vector of indices with > 0 elements is specified, R will return the elements of the vector in the order requested. For example, a[c(1,3,5)] and a[c(3,5,1)] will return the same values but in different orders.
a <- 11:20
# all elements returned
a[]
## [1] 11 12 13 14 15 16 17 18 19 20
# second element
a[2]
## [1] 12
# elements 3, 4, 5, 6, and 7
a[3:7]
## [1] 13 14 15 16 17
# error (not run)
# a[3:7,2]
# correct version of previous command
a[c(3:7,2)]
## [1] 13 14 15 16 17 12
# notice different ordering from previous command
a[c(2:7)]
## [1] 12 13 14 15 16 173.3.1.2 Selecting from a matrix
Matrices have 2 dimensions, so you must specify two dimension’s worth of indices. If you want all of one dimension (e.g., rows 1 to 3 and all columns), you can leave the spot for the index blank. Separate the dimensions using a comma. Positive indices select elements; negative elements remove elements.
I’ll reiterate this because it is a common mistake: you must specify all dimensions using a comma. Failing to do so will result in an error (bad) or unexpected results (worse).
Remember: in R, indices start at 1. In some languages such as Python indices start at 0.
a <- matrix(1:12, nrow=3, byrow=TRUE)
# value in row 1, column 2
a[1,2]
## [1] 2
# values in rows 1-3 and columns 1-2
a[1:3, 1:2]
## [,1] [,2]
## [1,] 1 2
## [2,] 5 6
## [3,] 9 10
# rows 1 and 2, all columns
a[1:2,]
## [,1] [,2] [,3] [,4]
## [1,] 1 2 3 4
## [2,] 5 6 7 8
# columns 1 and 2, all rows
a[,1:2]
## [,1] [,2]
## [1,] 1 2
## [2,] 5 6
## [3,] 9 10Negative indices remove elements, or return all positive indices.
Removing multiple elements with - can be a bit tricky. All elements of the vector of things to remove must be negative. In the first example below, R returns an error because you request elements -1, 0, 1, and 2. Element -1 does not exist, and you don’t want elements 1 and 2!
There are several correct ways to get rows other than 1 and 2. Notice that all of them result in indices with a 1-, a -2, and no positive values. The first option is probably the best for most situations.
Notice that all three of these lines return a vector, not a matrix. This is because a single row or column of a matrix simplifies to a vector. If you want to keep your result in matrix format, you can specify this using the argument drop=FALSE after the last dimension and a comma:
3.3.1.3 Selecting from a data frame
Selecting from a data frame using brackets is largely the same as selecting from a matrix. Just keep in mind that the result might come back as a vector (if you select rows in a single column) or a data frame (any other situation).
a <- iris[1:5,]
# returns a vector
a[,1]
## [1] 5.1 4.9 4.7 4.6 5.0
# returns a data frame
a[1,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
# returns a data frame
a[1:3,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
# returns a data frame
a[1:3, 1:3]
## Sepal.Length Sepal.Width Petal.Length
## 1 5.1 3.5 1.4
## 2 4.9 3.0 1.4
## 3 4.7 3.2 1.33.3.1.4 Selecting from arrays
Selecting from an array is a lot like selecting from a matrix, only with one more dimension. Consider an array with 3 dimensions: rows, columns, and “layers”38.
## , , 1
##
## [,1] [,2]
## [1,] 1 5
## [2,] 2 6
## [3,] 3 7
## [4,] 4 8
##
## , , 2
##
## [,1] [,2]
## [1,] 9 13
## [2,] 10 14
## [3,] 11 15
## [4,] 12 16
##
## , , 3
##
## [,1] [,2]
## [1,] 17 21
## [2,] 18 22
## [3,] 19 23
## [4,] 20 24Each element in any dimension matrix (2 dimensional array):
ar[1,,]
## [,1] [,2] [,3]
## [1,] 1 9 17
## [2,] 5 13 21
ar[,2,]
## [,1] [,2] [,3]
## [1,] 5 13 21
## [2,] 6 14 22
## [3,] 7 15 23
## [4,] 8 16 24
ar[,,3]
## [,1] [,2]
## [1,] 17 21
## [2,] 18 22
## [3,] 19 23
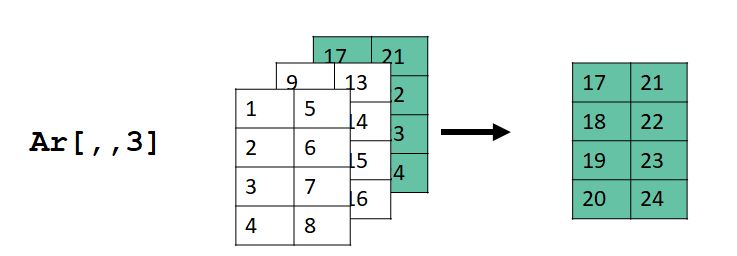
## [4,] 20 24The images below show what happened. For ar[1,,], the values in the first row (dimension 1), all columns (dimension 2), and all layers (dimension 3) were selected. Dimension 2 in the input ar became dimension 1 in the output, and dimension 3 in ar became dimension 2 in the output.
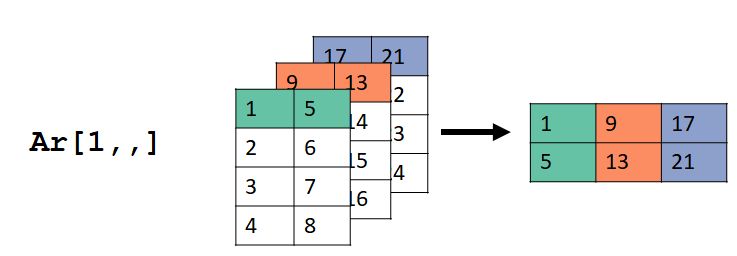
For ar[,2,], the values in the second column (dimension 2), all rows (dimension 1), and all layers (dimension 3) were selected. Dimension 2 in the input ar became dimension 1 in the output, and dimension 3 in ar became dimension 2 in the output.
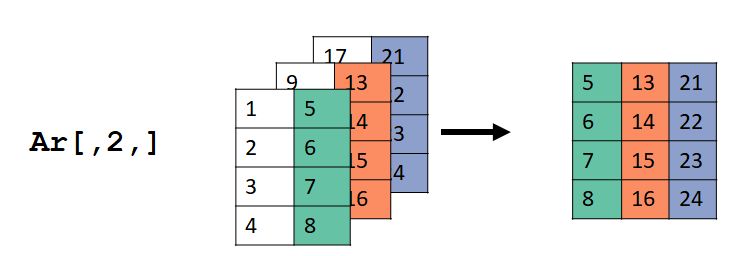
For ar[,,3], the values in the all rows (dimension 1), all columns (dimension 2), and layer 3 (dimension 3) were selected. Dimension 2 in the input ar became dimension 1 in the output, and dimension 3 in ar became dimension 2 in the output.
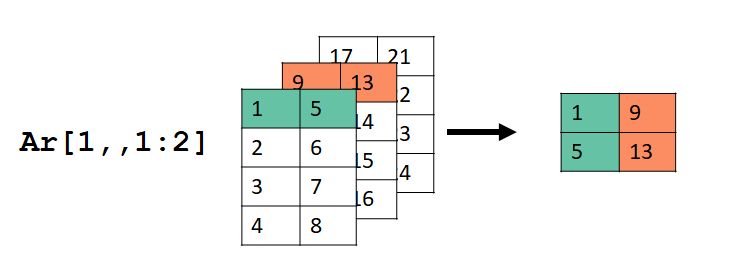
Selecting by \(\ge\) 1 dimension can be tricky. Consider this example:
Here we selected the first row (dimension 1), all columns (dimension 2), and layers 1 and 2 (dimension 3).
And if you want a headache, consider this example:
ar[1:2,,2:1]
## , , 1
##
## [,1] [,2]
## [1,] 9 13
## [2,] 10 14
##
## , , 2
##
## [,1] [,2]
## [1,] 1 5
## [2,] 2 6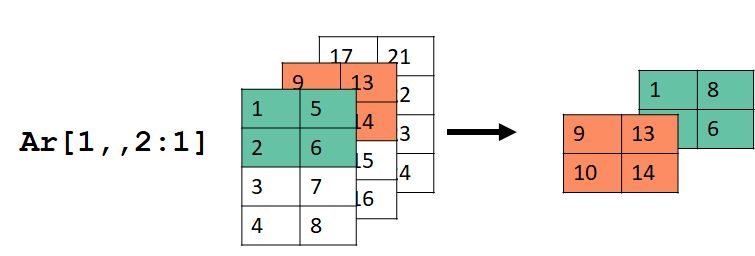
I can’t think of a reason why you would need to do something like the last example, but it’s nice to know that you could.
3.3.1.5 Selecting from lists
One of the most important kinds of R object is the list. A list can be thought of like a series of containers or buckets. Each bucket can contain completely unrelated things, or even other series of buckets. Each bucket is called an element of the list. Because of this flexibility, lists are extremely versatile and useful in R programming. Many function outputs are really lists (e.g., the outputs of lm() or t.test()).
Single elements of a list can be accessed with double brackets [[]]. Elements can be selected by name or by index. Elements that have a name can be selected by name using the $ symbol instead of by double brackets.
# make a list "a"
x <- runif(10)
y <- "zebra"
z <- matrix(1:12, nrow=3)
a <- list(e1=x, e2=y, e3=z)
a[[1]]
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695
a[[2]]
## [1] "zebra"
a[["e1"]]
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695
a$e1
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695Multiple elements of a list can be selected using single brackets. Doing so will return a new list containing the requested elements, not the requested elements themselves. This can be a little confusing. Compare the following two commands and notice that the first returns the 10 random numbers in the first element of a, while the second returns a list with 1 element, and that element is the numbers 1 through 10.
a[[1]]
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695
a[1]
## $e1
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695Next, observe what happens when we select more than one element of the list a. The output is a list.
a[1:2]
## $e1
## [1] 0.81537826 0.85151339 0.94974433 0.24860408 0.20883937 0.23072284
## [7] 0.80287954 0.06142657 0.65110133 0.21388695
##
## $e2
## [1] "zebra"
class(a[1:2])
## [1] "list"The reason for this syntax is that the single brackets notation allows you to extract multiple elements of the list at once to quickly make a new list.
3.3.2 Extracting and selecting data with logical tests
Selecting data from a data frame is one of the most common data manipulations. R offers several ways to accomplish this. One of the most fundamental is the function which(), returns the indices in a vector where a logical condition is TRUE. This means that you can select any piece of a dataset that can be identified logically. Consider the example below, which extracts the rows of the iris dataset where the variable Species matches the character string "setosa".
In most situations, which() can be omitted because it is implicit in the command. However, not using it can sometimes produce strange results. Because of this, I always use which() in my code. Including which() can also make your code a little clearer to someone who is not familiar with R. For example, the two commands below are exactly equivalent:
# not run (long output); try it on your machine!
iris[which(iris$Species == "setosa"),]
iris[iris$Species == "setosa",]What which() is really doing in the commands above is returning a vector of numbers. That vector is then used by the containing brackets [] to determine which pieces of the data frame to pull. The numbers returned by which() are the indices at which a logical vector is TRUE. Compare the results of the two commands below, which were used in the commands above to select from iris:
# logical vector:
iris$Species == "setosa"
## [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [13] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [25] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [37] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [49] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [61] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [73] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [85] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [97] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [109] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [121] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [133] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [145] FALSE FALSE FALSE FALSE FALSE FALSE
# which identifies which elements of vector are TRUE:
which(iris$Species == "setosa")
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
## [26] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50Multiple conditions can be used to select data using the & (and) and | (or) operators.
# not run (long output):
iris[which(iris$Species == "versicolor" & iris$Petal.Length <= 4),]
iris[which(iris$Species == "versicolor" | iris$Petal.Length <= 1.6),]The examples above could also be accomplished in several lines; in fact, splitting the selection criteria up can make for cleaner code. The commands below also show how splitting the selection commands up can help you abide by the “single point of definition” principle.
# not run (long output):
flag1 <- iris$Species == "versicolor"
flag2 <- iris$Petal.Length <= 4
iris[flag1 & flag2,]
iris[flag1 | flag2,]More than 2 conditions can be combined just as easily:
flag1 <- iris$Species == "versicolor"
flag2 <- iris$Petal.Length <= 4
flag3 <- iris$Petal.Length > 0.5
iris[flag1 & flag2 & flag3,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 54 5.5 2.3 4.0 1.3 versicolor
## 58 4.9 2.4 3.3 1.0 versicolor
## 60 5.2 2.7 3.9 1.4 versicolor
## 61 5.0 2.0 3.5 1.0 versicolor
## 63 6.0 2.2 4.0 1.0 versicolor
## 65 5.6 2.9 3.6 1.3 versicolor
## 70 5.6 2.5 3.9 1.1 versicolor
## 72 6.1 2.8 4.0 1.3 versicolor
## 80 5.7 2.6 3.5 1.0 versicolor
## 81 5.5 2.4 3.8 1.1 versicolor
## 82 5.5 2.4 3.7 1.0 versicolor
## 83 5.8 2.7 3.9 1.2 versicolor
## 90 5.5 2.5 4.0 1.3 versicolor
## 93 5.8 2.6 4.0 1.2 versicolor
## 94 5.0 2.3 3.3 1.0 versicolor
## 99 5.1 2.5 3.0 1.1 versicolorSelections can return 0 matches. This usually manifests as a data frame or matrix with 0 rows.
flag1 <- iris$Species == "versicolor"
flag2 <- iris$Petal.Length <= 1
iris[flag1 & flag2,]
## [1] Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <0 rows> (or 0-length row.names)A related function is the binary operator39 %in%. For a command like A &in% B, R returns a vector of logical values for each element in A, indicating whether those values occur in B.
## [1] FALSE FALSE TRUE TRUE TRUE## [1] TRUE TRUE TRUE FALSEOne example of how this might be useful is if you want to select observations from a dataset containing one of several values. The example below illustrates this with iris:
We’ll go over some more complicated selection methods in a future section, but what you’ve seen so far can be very powerful.
3.4 Managing dates and characters
Most of the data we deal with in R are numbers, but textual and temporal data are important two. Working with dates, times, and character strings can be significantly more complicated than working with numbers…so, I’ve put together an entire page on it. I am the first to admit that the approaches demonstrated here are a little “old-school” compared to some of the newer packages. In particular, many people prefer the tidyverse package lubridate (Grolemund and Wickham 2011) for temporal data. Much of this guide is based on Spector (2008).
3.4.1 Temporal data and dates
Temporal data can be read and interpreted by R in many formats. The general rule is to use the simplest way that you can, because the date and time classes can be very complicated. For example, don’t store date and time if you only need date. This doesn’t mean to discard potentially useful data; just to be judicious in what you import to and try to manipulate in R.
It should be noted that the easiest way to deal with dates and times is “don’t”. In many situations it’s actually not necessary to use temporal data classes. For example, if a field study was conducted in two years, and there is no temporal component to the question, it might be sufficient to just call the years year1 and year2 in the dataset. Or, if an elapsed time is used as a variable, it might make more sense to store the elapsed time (e.g., 10 minutes, 11.2 minutes, etc.) rather than use full date/time codes for each observation time.
3.4.1.1 Dates without times
The easiest way to handle dates is to use the base function as.Date(). This function converts character strings to the R date format. This implies that you will have a much easier time if you send dates to R in YYYY-MM-DD format. You can do this in Excel by setting a custom number format. Once in R, use as.Date() to convert your properly-formatted text strings into date objects. Once converted, R can do simple arithmetic such as finding intervals between dates.
# enter dates as text strings
a <- c("2021-07-23", "2021-08-01", "2021-09-03")
# convert to date
b <- as.Date(a)
# check classes
class(a)
## [1] "character"
class(b)
## [1] "Date"
# can do math with dates!
b[2] - b[1]
## Time difference of 9 daysThe examples above use the ISO 8601 date format, Year-Month-Day (or YYYY-MM-DD). This format has the advantage that sorting it alphanumerically is the same as sorting it chronologically. Each element has a fixed number of digits to avoid issues like January and October (months 1 and 10) being sorted next to each other.
I suggest that you get in the habit of storing dates this way. This format is the default date format in R (and many other programs), such that R can read dates in this format without additional arguments. If you insist on storing dates in other formats, you can still read those dates into R. You just need to tell as.Date() the format that the date is in. The syntax for formatting dates and times is handled by the function strptime() (see ?strptime for more examples). Something common to all of the formats below is that each element has a fixed number of digits, 4 for year and 2 for month or day. This is to avoid ambiguity: does “7/2/2019” “July second, 2019”, or “February 7, 2019”?. This also avoids issues arising from treating numbers as character strings. For example, January and October (months 1 and 10) might be sorted next to each other unless represented by “01” and “10” instead of “1” and “10”.
# YYYYMMDD
a <- "20210721"
as.Date(a, "%Y%m%d")
## [1] "2021-07-21"
# MMDDYYYY
a <- "07212021"
as.Date(a, "%m%d%Y")
## [1] "2021-07-21"
# MM/DD/YYYY
a <- "07/21/2021"
as.Date(a, "%m/%d/%Y")
## [1] "2021-07-21"
# DD-MM-YYYY
a <- "21-07-2021"
as.Date(a, "%d-%m-%Y")
## [1] "2021-07-21"An even simpler option for dates, if all dates are within the same year, is to use Julian dates (i.e., day-of-year). If you want to read Julian dates into R as dates, you can also use as.Date(). Notice in the example below that the Julian dates must be supplied as character strings with 3 digits, not as numerals. Notice also that R assigns the current year (2021) to the dates unless you specifically tell it to do something else.
a <- as.character(c(204, 213, 246))
as.Date(a, "%j")
## [1] "2024-07-22" "2024-07-31" "2024-09-02"
## not run:
## causes error because nchar("15") < 3:
#a <- c(15, 204, 213, 246)
#as.Date(a, "%j")
# convert all numbers to 3 digit
# character strings first with formatC():
a <- c(15, 204, 213, 246)
x <- formatC(a, width=3, flag="0")
as.Date(x, "%j")
## [1] "2024-01-15" "2024-07-22" "2024-07-31" "2024-09-02"Dates can be reformatted using the function format(). Notice that the output is a character string, not a date. The syntax for the conversion comes from strptime(), just as with as.Date() above. If you need to use a formatted date (character string) as a date (R class) you need to convert it with as.Date().
# make some dates as character strings and convert to date
a <- c("2021-07-23", "2021-08-01", "2021-09-03")
x <- as.Date(a)
# Julian day (day of year), as character
format(x, "%j")
## [1] "204" "213" "246"
# as number
as.numeric(format(x, "%j"))
## [1] 204 213 246
# ISO format
format(x, "%Y-%m-%d")
## [1] "2021-07-23" "2021-08-01" "2021-09-03"
# use month abbreviations, separate by slashes
format(x, "%b/%d/%y")
## [1] "Jul/23/21" "Aug/01/21" "Sep/03/21"3.4.1.2 Dates with times (and time zones)
Things get more complicated when time is included with a date, but the basic procedures are the same as date-only data. By default, R assumes that times are in the time zone of the current user. So, if you are collaborating with someone in a different time zone, you need to account for time zone in your code. Another default behavior is that R will assume that times without dates are on the current day.
The main function for working with dates and times is strptime(). This function takes in a character vector and a format argument, which tells R how the character vector is formatted. It returns objects of class POSIXlt and POSIXt, which contain date and time information in a standardized way.
# create some times as character strings
a <- c("01:15:03", "14:28:21")
# time with seconds
strptime(a, format="%H:%M:%S")
## [1] "2024-08-23 01:15:03 EDT" "2024-08-23 14:28:21 EDT"
# time with seconds dropped
strptime(a, format="%H:%M") # seconds dropped
## [1] "2024-08-23 01:15:00 EDT" "2024-08-23 14:28:00 EDT"Notice that the current date was included in the result of strptime() (at least, the date when I rendered this page with R Markdown).
Time zones should be specified when working with data that contain dates and times. Remember that R will assume the time is in the current user’s time zone. This almost always means the time zone used by the operating system in which R is running. If you did something silly like record data in one time zone and analyze it in another, you have to be careful. For example, I once ran a field study in Indiana where data were recorded in Eastern time. But, the office where I analyzed the data was in Missouri (Central time). R kept converting the time-stamped data to Central time, which interfered with the analysis until I learned how to work with time zones in R. To make things worse, data were recorded in the summer (Eastern daylight time) but analyzed in both early fall (Central daylight time) and late fall (Central standard time).
# make some date-times and convert to date-time class
a <- c("2021-07-23 13:21", "2021-08-01 09:15")
x <- strptime(a, "%Y-%m-%d %H:%M")
x
## [1] "2021-07-23 13:21:00 EDT" "2021-08-01 09:15:00 EDT"
# times without dates will be assigned current date:
a2 <- c("13:21", "09:15")
x2 <- strptime(a2, "%H:%M")
x2
## [1] "2024-08-23 13:21:00 EDT" "2024-08-23 09:15:00 EDT"Time zone names are not always portable across systems. R has a list of built-in time zones that it can handle, and these should work, but be careful and always validate any code that depends on time zones. The list of time zones R can use can be printed to the console with the command OlsonNames().
# US Central Daylight Time (5 hours behind UTC)
a <- c("2021-07-23 13:21", "2021-08-01 09:15")
# strptime() will assign whatever time zone you enter:
b <- strptime(a, "%Y-%m-%d %H:%M", tz="US/Eastern")
b
## [1] "2021-07-23 13:21:00 EDT" "2021-08-01 09:15:00 EDT"
b <- strptime(a, "%Y-%m-%d %H:%M", tz="US/Central")
b
## [1] "2021-07-23 13:21:00 CDT" "2021-08-01 09:15:00 CDT"Internally, dates and times are stored as the number of seconds elapsed since midnight (00:00 hours) on 1 January 1970, in UTC. You can see these values with as.numeric(). The time zone of a value affects what is printed to the console. That time zone is stored by R as an attribute of the value. Attributes are a kind of metadata associated with some R data types that can be accessed with attributes().
## [1] "US/Central" "CST" "CDT"To change the time zone associated with value, simply set the attribute to the desired time zone. This is basically telling R the time zone that a date and time refer to. Note that will change the actual value of a date/time. It does NOT translate times in one time zone to another.
as.numeric(b)
## [1] 1627064460 1627827300
attr(b, "tzone") <- "America/Los_Angeles"
# different by 7200 seconds (2 hours)
as.numeric(b)
## [1] 1627071660 1627834500To convert one time zone to another, the values must first be converted to the POSIXct class.
b <- strptime(a, "%Y-%m-%d %H:%M", tz="US/Central")
b2 <- as.POSIXct(b)
# check time zone
attributes(b2)$tzone
## [1] "US/Central"
# check actual time (seconds since epoch)
as.numeric(b2)
## [1] 1627064460 1627827300
# change time zone of POSIXct object
attr(b2, "tzone") <- "Europe/London"
# actual time same as before:
as.numeric(b2)
## [1] 1627064460 16278273003.4.1.3 Working with dates and times
Why should we go through all of this trouble to format dates and times as dates and times? The answer is that R can perform many useful calculations with dates and times. One of these is time intervals by using subtraction or the function difftime(). Subtracting two date/times will return an answer but perhaps not in the units you want; use difftime() if you want to specify the units.
# how many days, hours, and minutes until Dr. Green can retire?
## current time:
a <- Sys.time()
## retirement date per USG:
b <- "2048-07-01 00:00"
## convert to POSIXlt
x <- strptime(a, "%Y-%m-%d %H:%M")
y <- strptime(b, "%Y-%m-%d %H:%M")
## calculate time in different units:
difftime(y,x, units="days")
## Time difference of 8712.451 days
difftime(y,x, units="hours")
## Time difference of 209098.8 hours
difftime(y,x, units="mins")
## Time difference of 12545930 mins
y-x
## Time difference of 8712.451 daysAnother application: what is the time difference between Chicago and Honolulu?
a <- c("2021-07-23 13:21")
x <- strptime(a, "%Y-%m-%d %H:%M", tz="US/Central")
y <- strptime(a, "%Y-%m-%d %H:%M", tz="US/Hawaii")
difftime(x,y)## Time difference of -5 hoursOddly, difftime() cannot return years. This is probably because there are different types of years (sidereal, tropical, anomalistic, lunar, etc.). You can convert a time difference in days to one in years by dividing by the number of days in the kind of year you want. The example below converts to does this and changes the “units” attribute of the difftime output to match.
d <- difftime(y,x, units="days")
# julian year
d <- d/365.25
attr(d, "units") <- "years"
d
## Time difference of 0.0005703856 yearsThe Julian year is useful because it is defined in terms of an SI units as 31557600 seconds, or 365.25 days40. Most of the alternative years at the link above vary by less than a few minutes.
R can also generate regular sequences of dates and times just like it can numeric values. See ?seq.date for details and syntax.
a <- as.Date("2021-07-23")
seq(a, by="day", length=10)
## [1] "2021-07-23" "2021-07-24" "2021-07-25" "2021-07-26" "2021-07-27"
## [6] "2021-07-28" "2021-07-29" "2021-07-30" "2021-07-31" "2021-08-01"
seq(a, by="days", length=6)
## [1] "2021-07-23" "2021-07-24" "2021-07-25" "2021-07-26" "2021-07-27"
## [6] "2021-07-28"
seq(a, by="week", length=10)
## [1] "2021-07-23" "2021-07-30" "2021-08-06" "2021-08-13" "2021-08-20"
## [6] "2021-08-27" "2021-09-03" "2021-09-10" "2021-09-17" "2021-09-24"
seq(a, by="6 weeks", length=10)
## [1] "2021-07-23" "2021-09-03" "2021-10-15" "2021-11-26" "2022-01-07"
## [6] "2022-02-18" "2022-04-01" "2022-05-13" "2022-06-24" "2022-08-05"
seq(a, by="-4 weeks", length=5)
## [1] "2021-07-23" "2021-06-25" "2021-05-28" "2021-04-30" "2021-04-02"3.4.2 Character data (text)
Recall from earlier that “character” is just another type of data in R–the type that contains text. This means that text values can be stored and manipulated in many of the same ways as numeric data. The vectorized functions in R can make managing your text-based data very fast and efficient.
A value containing text, or stored as text, is called a character string. Character strings can be stored in many R objects. A set of character strings is called a character vector.
3.4.2.1 Combining character strings
Character strings can be combined together into larger character strings using the function paste(). Notice that this is not the same as making a vector containing several character strings. That operation is done with c().
x <- paste("a", "b", "c")
y <- c("a", "b", "c")
x
## [1] "a b c"
y
## [1] "a" "b" "c"
length(x)
## [1] 1
length(y)
## [1] 3The function paste(), and its cousin paste0(), combine their arguments together and output a character string. The inputs do not have to be character strings, but the outputs will be. The difference between paste() and paste0() is that paste0() does not put any characters between its inputs, while paste() can place any valid character between its inputs. The default separator is a space.
paste0("a", "zebra", 3)
## [1] "azebra3"
paste("a", "zebra", 3)
## [1] "a zebra 3"
paste("a", "zebra", 3, sep="_")
## [1] "a_zebra_3"
paste("a", "zebra", 3, sep=".")
## [1] "a.zebra.3"
paste("a", "zebra", 3, sep="/")
## [1] "a/zebra/3"
paste("a", "zebra", 3, sep="giraffe")
## [1] "agiraffezebragiraffe3"One of my favorite uses for the paste functions is to automatically generate folder names and output file names. The example below adds the current date (Sys.Date()) into a file name.
# not run
top.dir <- "C:/_data"
out.dir <- paste(top.dir, "test", sep="/")
out.name <- paste0("output_", Sys.Date(), ".csv")
write.csv(iris, paste(geo.dir, out.name, sep="/"))When the inputs to a paste() function are contained in a single vector, then the desired separator must be supplied using the argument collapse instead of sep.
## [1] "a-b-c"3.4.2.2 Printing character strings
The cat() function can not only put strings together, but also print them to the R console. This is useful if you want to generate messages from inside functions or loops. When cat() receives inputs that are not character strings, it evaluates them and converts them to character strings. The examples below show how inputs can be evaluated and printed. It also illustrates the use of \n and \t to insert new lines and tabs, respectively. Finally, the command flush.console() ensures that the console is updated with the most recent outputs.
# using cat() in a function:
my.fun <- function(x){
cat("You entered", x, "!", "\n")
cat("\t", "1 more is", x+1, "\n")
}
my.fun(7)## You entered 7 !
## 1 more is 8## You entered 42 !
## 1 more is 43# using cat() in a loop:
for(i in 1:5){
cat("starting iteration", i, "\n")
for(j in 1:3){
cat("\t", "iteration", i, "part", j, "complete!", "\n")
flush.console()
}
cat("iteration", i, "complete!", "\n")
}## starting iteration 1
## iteration 1 part 1 complete!
## iteration 1 part 2 complete!
## iteration 1 part 3 complete!
## iteration 1 complete!
## starting iteration 2
## iteration 2 part 1 complete!
## iteration 2 part 2 complete!
## iteration 2 part 3 complete!
## iteration 2 complete!
## starting iteration 3
## iteration 3 part 1 complete!
## iteration 3 part 2 complete!
## iteration 3 part 3 complete!
## iteration 3 complete!
## starting iteration 4
## iteration 4 part 1 complete!
## iteration 4 part 2 complete!
## iteration 4 part 3 complete!
## iteration 4 complete!
## starting iteration 5
## iteration 5 part 1 complete!
## iteration 5 part 2 complete!
## iteration 5 part 3 complete!
## iteration 5 complete!The other printing function, sink(), is specifically for printing outputs into text files. Refer back to the section on data import/export for an example.
3.4.2.3 Matching character strings
Like numeric data, character strings can be matched exactly with the comparison operator ==.
## [1] TRUE FALSE## [1] 1## [1] 2## [1] TRUE## [1] FALSENotice that this kind of matching is case-sensitive. If you need to match regardless of case, you can either force all of the comparands to be the same case, or use regular expressions (see next section).
R is smart enough to sort text alphabetically, and to make comparisons based on alphabetical sorting.
3.4.2.4 Searching with grep() and grepl()
Regular expressions, or regex, are a syntax for searching text strings. Learning how to use regular expressions is easily a semester-long course on its own. Here we’ll just explore a few common use cases. The R functions that deal with regular expressions are grep() and its cousins. The most commonly used versions are grep() and grepl().
grep()returns the indices in the input vector that match the pattern. The length of the output will be the number of matches.grepl()returns a logical value (TRUE or FALSE) telling whether each element in the input vector matches the pattern. The length of the output will be same as the length of the input.
This means that grepl() is analogous to using == to test for equality, while grep() is analogous to using which(). Compare the results of the two commands on the simple vector x below. It’s not hard to imagine how powerful these functions can be.
x <- c("zebra", "hippo", "lion", "tiger")
grep("e", x)
## [1] 1 4
grepl("e", x)
## [1] TRUE FALSE FALSE TRUESome of the arguments to grep() can save you some time. Setting value=TRUE will return the values where the input matches the pattern.
grep("e", x, value=TRUE)
## [1] "zebra" "tiger"
# equivalent to:
x[grep("e", x)]
## [1] "zebra" "tiger"Setting invert=TRUE will find elements of the input that do NOT match the pattern.
By default, grep() is case-sensitive. This can be overridden with the option ignore.case.
x <- c("zebra", "hippo", "lion", "tiger", "Tiger")
grep("tiger", x)
## [1] 4
grep("Tiger", x)
## [1] 5
grep("tiger", x, ignore.case=TRUE)
## [1] 4 5
grep("TIGER", x, ignore.case=TRUE)
## [1] 4 5We will see some uses of grep() and its cousins in the examples later.
3.4.2.5 Replacing parts of character strings
Text within character strings can be replaced or modified in several ways. If you are comfortable with regular expressions (see previous section) then you can use function gsub(). The syntax for gsub() is similar to that of grep(), with an additional input for the replacement string. Note that gsub() will replace every occurrence of the pattern, whether or not an occurrence is a whole word or not. The related function sub() works like gsub(), but only replaces the first occurrence of the pattern.
x <- c("zebra", "hippo", "lion", "tiger", "Tiger")
gsub("tiger", "giraffe", x)
## [1] "zebra" "hippo" "lion" "giraffe" "Tiger"
gsub("tiger", "giraffe", x, ignore.case=TRUE)
## [1] "zebra" "hippo" "lion" "giraffe" "giraffe"
gsub("z", "Z", x)
## [1] "Zebra" "hippo" "lion" "tiger" "Tiger"
sub("tiger", "giraffe", x, ignore.case=TRUE)
## [1] "zebra" "hippo" "lion" "giraffe" "giraffe"3.4.2.6 Extracting from and splitting character strings
Character strings can be split up using the function strsplit() (short for “string split”). This function can use regular expressions like grep() or gsub() can. It works by breaking a character string apart at a given text string. Unfortunately, its output can be a little confusing if you’re not used to working with R objects:
## [[1]]
## [1] "Peromyscus" "leucopus"
##
## [[2]]
## [1] "Sigmodon" "hispidus"
##
## [[3]]
## [1] "Microtus" "pennsylvanicus"Function strsplit() returns a list with one element for each element in the input vector. Each element of that list is a character vector, consisting of the pieces of that element of the input vector split by the splitting string. So, if you want only one part of each element of the input vector, you need to extract them from the output list. In this example, we can get the generic epithets by pulling the first element of each list element. The function sapply() applies a function to each element of a list and returns a result with the simplest possible structure (sapply is short for “apply and simplify”). The two commands below are equivalent, but the first is much simpler and should be the method you use.
sapply(strsplit(x, "_"), "[", 1)
## [1] "Peromyscus" "Sigmodon" "Microtus"
# equivalent, not as easy:
do.call(c, lapply(strsplit(x, "_"), "[", 1))
## [1] "Peromyscus" "Sigmodon" "Microtus"The “sapply-strsplit” method is useful for breaking apart strings whose parts carry information. One common use case is breaking up dates into year, month, and day. Note that there are more elegant ways to do this using the R date classes, but treating dates as text strings can be convenient because the date and time classes are such a pain in the neck. When using this method, you will need to keep track of when date components are stored as text and when they are stored as numbers.
The code block below shows how to break apart dates using strsplit() and sapply().
x <- c("2019-08-03", "2020-08-15", "2021-08-07")
ssx <- strsplit(x, "-")
a <- data.frame(year=sapply(ssx, "[", 1),
mon=sapply(ssx, "[", 2),
day=sapply(ssx, "[", 3))
class(a$year) # character, not a number!
## [1] "character"
class(a$mon) # character, not a number!
## [1] "character"
class(a$day) # character, not a number!
## [1] "character"
a$year <- as.numeric(a$year)
a$mon <- as.numeric(a$mon)
a$day <- as.numeric(a$day)
class(a$year) # numbers!
## [1] "numeric"
class(a$mon) # numbers!
## [1] "numeric"
class(a$day) # numbers!
## [1] "numeric"3.4.2.7 Matching text strings
As biologists we often keep our data in several separate spreadsheets or tables. These tables are interrelated by a set of “key” variables. If that sounds like a sketchy, haphazard way to build a database, that’s because it is. Databases are outside the scope of this course, but we will learn how to relate tables using R. If you’ve ever used the VLOOKUP function in Excel, then you know exactly what problem match() is designed to solve.
The function match() finds the indices of the matches of its first argument in its second. In other words, it searches for the values in one vector in a second vector, then returns the positions of the matches from the second vector. A simple example is below:
vec1 <- c("gene1", "gene2", "gene3", "gene4")
vec2 <- c("gene1", "gene2", "gene1", "gene2",
"gene5", "gene3", "gene3", "gene4")
vec3 <- c("gene1", "gene2", "gene1", "gene2",
"gene4", "gene3", "gene3", "gene4")
match(vec1, vec2)
## [1] 1 2 6 8
match(vec2, vec1)
## [1] 1 2 1 2 NA 3 3 4
match(vec3, vec1)
## [1] 1 2 1 2 4 3 3 4The first example shows what match() is doing. For element in the first vector, match() returns the first position where that value occurs in the second vector. The second example shows what happens when trying to match a value that is not in the second vector: R returns NA. The third example shows how match() can be used to relate tables. The longer vector vec3 is matched to the shorter vector vec1, which contains each unique value only once. If vec1 was a variable in a data frame that contained other variables associated with genes, then the result of match(vec3, vec1) would return the rows in vec1’s data frame associated with each row of vec3’s data frame.
The code below demonstrates how match() might be used in practice. This example uses three tables from a real dataset. The three tables contain data and metadata for samples taken from stream across Georgia. The commands use match() to assign site numbers, dates, and geographic coordinates to observations using different key variables. We’ll revisit these data again in the next section where we see how to merge datasets.
# not run (see next section)
options(stringsAsFactors=FALSE)
name1 <- "dat_samples_2021-08-04.csv"
name2 <- "dat_stream_indices_2021-08-04.csv"
name3 <- "loc_meta_vaa_2020-11-18.csv"
x1 <- read.csv(name1, header=TRUE)
x2 <- read.csv(name2, header=TRUE)
x3 <- read.csv(name3, header=TRUE)
# x2 contains the response variable
dat <- x2
# add site name, year, and date from x1
matchx <- match(dat$uocc, x1$uocc)
dat$site <- x1$loc[matchx]
dat$year <- x1$year[matchx]
dat$doy <- x1$doy[matchx]
# equivalent to:
#dat$site <- x1$loc[match(dat$uocc, x1$uocc)]
#dat$year <- x1$year[match(dat$uocc, x1$uocc)]
#dat$doy <- x1$doy[match(dat$uocc, x1$uocc)]
# grab latitude and longitude from x3
matchx <- match(dat$site, x3$loc)
dat$lat <- x3$lat[matchx]
dat$long <- x3$long[matchx]3.4.2.8 Standardizing numbers as text
The function formatC() formats numbers in a manner similar to the language C. This is useful for naming objects or parts of objects with numeric names that have a fixed width or format. The most common use case is converting numbers like “1, 12, 112” to strings like “001, 012, 112”. The example below uses formatC() in combination with paste() to generate some file names.
file.nums <- formatC(1:4, width=3, flag="0")
file.names <- paste("file", file.nums, sep="_")
file.names## [1] "file_001" "file_002" "file_003" "file_004"This is useful for generating text strings that need to be sorted. In the example above, the files “file_001.txt”, “file_002.txt”, “file_003.txt”, and “file_004.txt” would be in the order you expect, and a file like “file_010.txt” would come after “file009.txt”. If the leading zeros had not been added, then the order would be “file_1.txt”, “file_10.txt”, “file_2.txt”, “file_3.txt”, etc. (notice that 10 was between 1 and 2).
3.5 Data frame management
Most of the time when you work with data in R, those data will be stored in data frames. A data frame looks like a matrix, but functions in many ways spreadsheet. Internally, a data frame is really a list, but for most day-to-day use you can think of a data frame as a table.
Being able to manage data frames is an important part of working with R. This page demonstrates some of the most common data frame operations.
3.5.1 Data frame structure
Data frames and matrices look very similar, but their internal structures are very different. This has huge implications for how you work with and manipulate data frames and matrices. The figure below shows some of the most important differences.
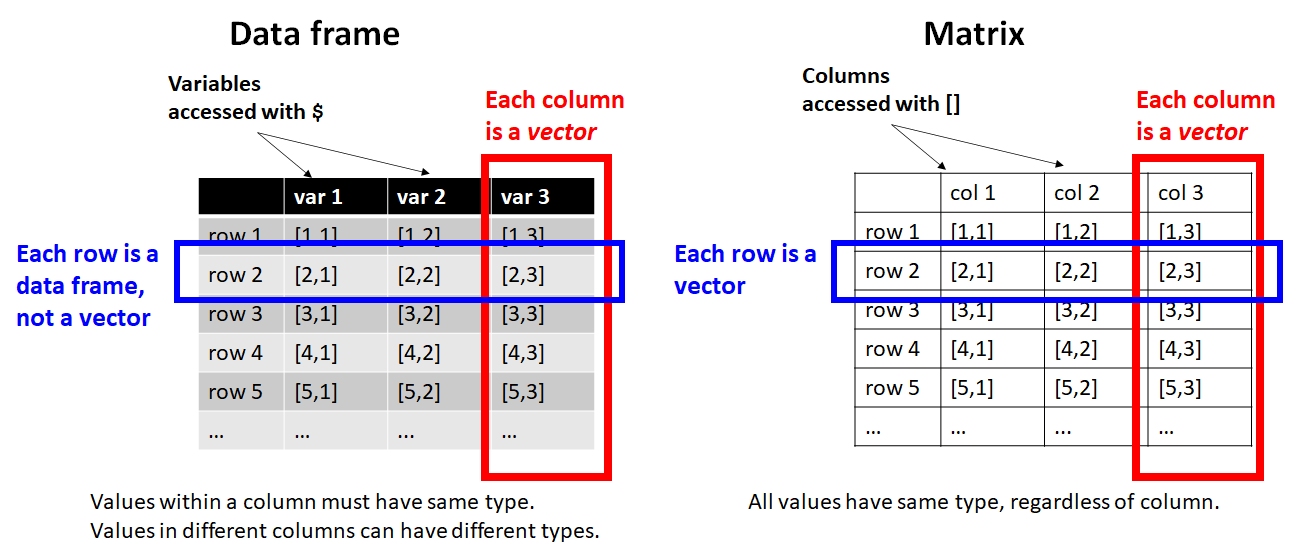
All pieces of a matrix are accessed using bracket notation ([]). Pieces of a data frame can also be accessed using brackets, but the dollar sign $ is usually used to access columns.
# single variables (columns): $
iris$Petal.Length
## [1] 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 1.5 1.6 1.4 1.1 1.2 1.5 1.3 1.4
## [19] 1.7 1.5 1.7 1.5 1.0 1.7 1.9 1.6 1.6 1.5 1.4 1.6 1.6 1.5 1.5 1.4 1.5 1.2
## [37] 1.3 1.4 1.3 1.5 1.3 1.3 1.3 1.6 1.9 1.4 1.6 1.4 1.5 1.4 4.7 4.5 4.9 4.0
## [55] 4.6 4.5 4.7 3.3 4.6 3.9 3.5 4.2 4.0 4.7 3.6 4.4 4.5 4.1 4.5 3.9 4.8 4.0
## [73] 4.9 4.7 4.3 4.4 4.8 5.0 4.5 3.5 3.8 3.7 3.9 5.1 4.5 4.5 4.7 4.4 4.1 4.0
## [91] 4.4 4.6 4.0 3.3 4.2 4.2 4.2 4.3 3.0 4.1 6.0 5.1 5.9 5.6 5.8 6.6 4.5 6.3
## [109] 5.8 6.1 5.1 5.3 5.5 5.0 5.1 5.3 5.5 6.7 6.9 5.0 5.7 4.9 6.7 4.9 5.7 6.0
## [127] 4.8 4.9 5.6 5.8 6.1 6.4 5.6 5.1 5.6 6.1 5.6 5.5 4.8 5.4 5.6 5.1 5.1 5.9
## [145] 5.7 5.2 5.0 5.2 5.4 5.1
# multiple variables: brackets []
## head() shows only first few rows (output very long)
a <- iris[,c("Petal.Length", "Sepal.Length")]
head(a)
## Petal.Length Sepal.Length
## 1 1.4 5.1
## 2 1.4 4.9
## 3 1.3 4.7
## 4 1.5 4.6
## 5 1.4 5.0
## 6 1.7 5.4
# rows: brackets []
# remember the comma!
iris[1:4,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
iris[1,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosaNotice that the syntax for accessing a data frame with [] is the same as for accessing a matrix.
When a data frame is printed to the console, this is only a representation of the data frame’s true form: a list. This is why single columns are accessible by a $: this is the symbol for accessing an element of a list. The list nature of a data frame is also why rows of a dataset are actually data frames themselves and not vectors.
3.5.2 Common data frame operations
3.5.2.1 Adding and deleting variables
New variables can be added to a data frame by assignment (<-). Assigning values to a variable in a data frame will create that variable if it doesn’t already exist–and overwrite values if the variable is already in the data frame.
x <- iris[1:5,] # spare copy for demonstration
x$Petal.Area <- apply(x[,3:4], 1, prod)
# shows new variable:
x## Sepal.Length Sepal.Width Petal.Length Petal.Width Species Petal.Area
## 1 5.1 3.5 1.4 0.2 setosa 0.28
## 2 4.9 3.0 1.4 0.2 setosa 0.28
## 3 4.7 3.2 1.3 0.2 setosa 0.26
## 4 4.6 3.1 1.5 0.2 setosa 0.30
## 5 5.0 3.6 1.4 0.2 setosa 0.28## Sepal.Length Sepal.Width Petal.Length Petal.Width Species Petal.Area
## 1 5.1 3.5 1.4 0.2 setosa 3
## 2 4.9 3.0 1.4 0.2 setosa 3
## 3 4.7 3.2 1.3 0.2 setosa 3
## 4 4.6 3.1 1.5 0.2 setosa 3
## 5 5.0 3.6 1.4 0.2 setosa 3Variables can also be added using function cbind(). This name is short for “column bind”41. To use cbind(), the input data frames must have the same number of rows. Note that cbind() sticks the data frames together as they are. If the records in the inputs need to be matched up and are not already in the same order, then either the data frames need to be aligned, merged with function merge(), or the variables need to be added using match() and a key variable.
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.5 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.4 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosaVariables can be deleted by setting them to NULL (notice all caps) or by making a new version of the data frame without the variable. If I am only removing a few variables, I prefer setting them individually to NULL. This makes it less likely that I will accidentally remove the wrong columns.
x$Petal.Area <- NULL
x
## Sepal.Length Sepal.Width
## 1 5.1 3.5
## 2 4.9 3.0
## 3 4.7 3.2
## 4 4.6 3.1
## 5 5.0 3.6
# delete 1 variable
x <- x[,which(names(x) != "Petal.Area")]
x
## Sepal.Length Sepal.Width
## 1 5.1 3.5
## 2 4.9 3.0
## 3 4.7 3.2
## 4 4.6 3.1
## 5 5.0 3.6
# delete >1 variable
x <- x[,which(!names(x) %in% c("Petal.Area", "Petal.Length"))]
x
## Sepal.Length Sepal.Width
## 1 5.1 3.5
## 2 4.9 3.0
## 3 4.7 3.2
## 4 4.6 3.1
## 5 5.0 3.63.5.2.2 Modifying variables
Variables can be modified by assigning them their new values. Note that this overwrites the previous values. If you think you might want the old values for some reason, save the modified values in a new variable instead.
3.5.2.3 Changing values in a variable using a rule
By combining the methods for data selection and assignment, values in a variable can be replaced according to their values.
# not run (long output), but try it on your machine!
x <- iris
# select and replace by one condition:
flag <- which(x$Sepal.Length >= 6)
x$Sepal.Length[flag] <- 5.99
x
# select and replace by 2 conditions:
flag1 <- which(x$Petal.Length >= 3)
flag2 <- which(x$Petal.Length <= 5)
x$Petal.Length[intersect(flag1, flag2)] <- 4
x3.5.2.4 Renaming variables
Renaming variables in Excel is easy: just enter the new name into the cell at the top of a column. Things are a little more complicated in R but not much.
One way is to use the data frame editor. Use the edit() function to open the data editor, a spreadsheet-like interface for working with data frames. Then click on the column name and enter the new name (and then press ENTER). You can also edit individual values this way, but I don’t recommend this method for routine use because there is no record in your script of what was done.
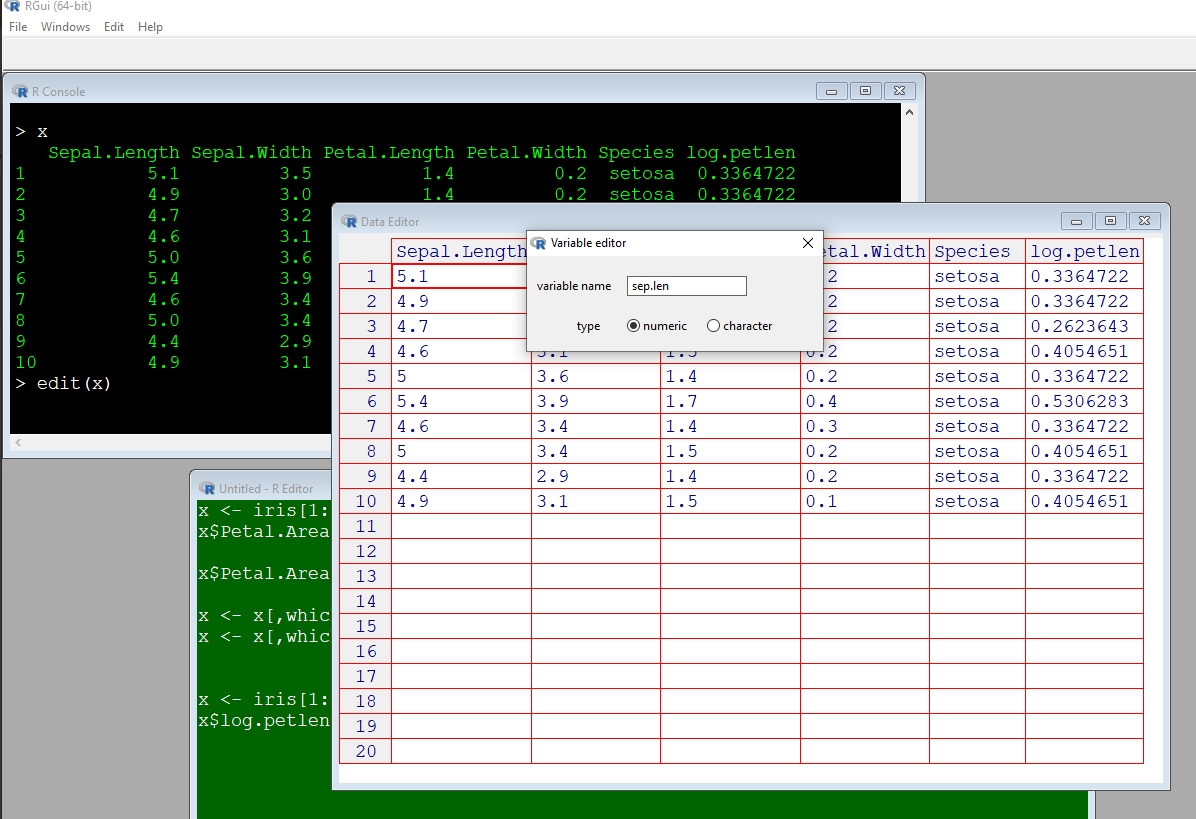
The better way is to rename the variable in code. There are a few ways to do this, but they all involve accessing the variable names using function names().
x <- iris[1:5,] # spare copy for demonstration
# by column number:
names(x)[1] <- "sep.len"
x
## sep.len Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
# match name exactly:
names(x)[which(names(x) == "Petal.Length")] <- "pet.len"
x
## sep.len Sepal.Width pet.len Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
# by position:
names(x)[ncol(x)] <- "species"
x
## sep.len Sepal.Width pet.len Petal.Width species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
names(x)[1] <- "petal.len"
x
## petal.len Sepal.Width pet.len Petal.Width species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
# change several names at once:
names(x)[1:4] <- paste0("x", 1:4)
x
## x1 x2 x3 x4 species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosaFinally, you can make a new copy of the variable with the name you want, then delete the original version. This has the disadvantage of changing the order of columns.
3.5.2.5 Rearranging columns
Reordering records or columns within a data frame is accomplished by creating a new version of the dataset with the desired arrangement. This rearrangement is accomplished using methods we have already seen.
x <- iris[1:5,]
# rearrange by number
x <- x[,c(5, 3, 4, 1, 2)]
x
## Species Petal.Length Petal.Width Sepal.Length Sepal.Width
## 1 setosa 1.4 0.2 5.1 3.5
## 2 setosa 1.4 0.2 4.9 3.0
## 3 setosa 1.3 0.2 4.7 3.2
## 4 setosa 1.5 0.2 4.6 3.1
## 5 setosa 1.4 0.2 5.0 3.6
# rearrange by name
col.order <- c("Species", "Sepal.Length", "Sepal.Width",
"Petal.Length", "Petal.Width")
x <- x[,col.order]
x
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 setosa 5.1 3.5 1.4 0.2
## 2 setosa 4.9 3.0 1.4 0.2
## 3 setosa 4.7 3.2 1.3 0.2
## 4 setosa 4.6 3.1 1.5 0.2
## 5 setosa 5.0 3.6 1.4 0.23.5.2.6 Sorting and ordering data
Sorting data frames is mostly done using the function order(). This function is usually put inside brackets in the rows slot. The arguments to order() define the variables by which the data frame will be sorted. Rows are sorted by the first variable, then the second, and so on. By default, rows are placed in ascending order.
# not run (long output), but try on your machine!
## order by conc (ascending)
DNase
DNase[order(DNase$conc),]
## order by species (alphabetical ascending),
## then by petal length (ascending)
iris
iris[order(iris$Species, iris$Petal.Length),]You can multiple variables to order(), by name, and R will order the rows in each variable in the order supplied. By default, records are sorted in ascending order. If you want a mix of ascending and descending order, you have to specify the radix method and specify whether you want the order to be decreasing (TRUE) or not (FALSE) for each variable:
3.5.2.7 Adding or removing rows
Removing rows works a lot like removing columns. You can use the bracket notation to either remove the rows with negative indices, or make a new version of the data frame with only some of the rows.
x <- iris[1:10,]
# remove by row number
x[-1,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
x[-c(1:3),]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
x[-c(1, 3, 5, 7, 9),]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 2 4.9 3.0 1.4 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
# or, make a new copy with the
# rows to keep
x[2:10,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
x[c(2, 4, 6, 8, 10),]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 2 4.9 3.0 1.4 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosaAdding rows can be done with rbind() (short for “row bind”). This function is analogous to cbind(), which joins objects by their columns. Adding rows to a data frame is tantamount to combining it with another data frame. The data frames being combined must have the same columns, with the same names, but the order of columns can be different.
x <- iris[1:10,]
y <- iris[15:20,]
# columns match exactly:
rbind(x, y)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## 15 5.8 4.0 1.2 0.2 setosa
## 16 5.7 4.4 1.5 0.4 setosa
## 17 5.4 3.9 1.3 0.4 setosa
## 18 5.1 3.5 1.4 0.3 setosa
## 19 5.7 3.8 1.7 0.3 setosa
## 20 5.1 3.8 1.5 0.3 setosa
# columns in different order:
y2 <- y[,c(3,5,2,1,4)]
rbind(x,y2)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## 15 5.8 4.0 1.2 0.2 setosa
## 16 5.7 4.4 1.5 0.4 setosa
## 17 5.4 3.9 1.3 0.4 setosa
## 18 5.1 3.5 1.4 0.3 setosa
## 19 5.7 3.8 1.7 0.3 setosa
## 20 5.1 3.8 1.5 0.3 setosa
# result inherits order of columns from first argument:
rbind(y2,x)
## Petal.Length Species Sepal.Width Sepal.Length Petal.Width
## 15 1.2 setosa 4.0 5.8 0.2
## 16 1.5 setosa 4.4 5.7 0.4
## 17 1.3 setosa 3.9 5.4 0.4
## 18 1.4 setosa 3.5 5.1 0.3
## 19 1.7 setosa 3.8 5.7 0.3
## 20 1.5 setosa 3.8 5.1 0.3
## 1 1.4 setosa 3.5 5.1 0.2
## 2 1.4 setosa 3.0 4.9 0.2
## 3 1.3 setosa 3.2 4.7 0.2
## 4 1.5 setosa 3.1 4.6 0.2
## 5 1.4 setosa 3.6 5.0 0.2
## 6 1.7 setosa 3.9 5.4 0.4
## 7 1.4 setosa 3.4 4.6 0.3
## 8 1.5 setosa 3.4 5.0 0.2
## 9 1.4 setosa 2.9 4.4 0.2
## 10 1.5 setosa 3.1 4.9 0.13.5.3 Other data frame operations
3.5.3.1 Merging datasets
Data sets for a complex project might reside in several tables, related by key variables. These are variables that are shared by more than one table, and define which records in each table are related to records in another table. This is exactly what relational databases do, but most biologists have small enough datasets that databases aren’t necessary.
Data frames can be combined by manually matching records between tables, using the function match(). The example below uses three interrelated tables from a stream dataset I work with. You need the datasets dat_samples_2021-08-04.csv, dat_stream_indices_2021-08-04.csv, and loc_meta_vaa_2020-11-18.csv. Download them and put them in your R home directory.
options(stringsAsFactors=FALSE)
# data file names
name1 <- "dat_samples_2021-08-04.csv"
name2 <- "dat_stream_indices_2021-08-04.csv"
name3 <- "loc_meta_vaa_2020-11-18.csv"
# import
x1 <- read.csv(name1, header=TRUE)
x2 <- read.csv(name2, header=TRUE)
x3 <- read.csv(name3, header=TRUE)Notice that the tables have only “key” variables in common, and tables don’t necessarily share any variables with every other table.
intersect(names(x1), names(x2))
## [1] "uocc"
intersect(names(x1), names(x3))
## [1] "loc"
intersect(names(x2), names(x3))
## character(0)Table x2 contains the response variable, so we’ll use it to build main dataset dat.
dat <- x2
# add site name, year, and date from x1
# by matching unique occasion (uocc) from
# dat to metadata table x1
matchx <- match(dat$uocc, x1$uocc)
dat$site <- x1$loc[matchx]
dat$year <- x1$year[matchx]
dat$doy <- x1$doy[matchx]
# dat now has site, which allows it to be
# related to the geographic metadata x3.
# grab latitude and longitude from x3
matchx <- match(dat$site, x3$loc)
dat$lat <- x3$lat[matchx]
dat$long <- x3$long[matchx]
# inspect the results
head(dat)
## uocc ind site year doy lat long
## 1 occ_001 48.40576 RV_01_16345 2017 54 34.05400 -83.24100
## 2 occ_002 56.78960 RV_01_240 2006 25 32.67461 -81.46906
## 3 occ_003 60.06336 RV_01_241 2003 33 33.58468 -82.65235
## 4 occ_004 21.30232 RV_01_242 2002 292 33.38387 -82.00422
## 5 occ_005 51.50136 RV_01_243 2002 12 33.83757 -82.91251
## 6 occ_006 73.91343 RV_01_244 2007 16 34.95895 -83.57158Matching many variables by hand can become very tedious and error-prone. An alternative, and usually faster way is the function merge(). This is a shortcut for the matching that we did manually above. The downside of using merge() is that it can be a little obtuse–merge() will attempt to match records exactly, which may or may not return the results you expect. Using merge() can also sometimes result in many duplicate columns or extra rows in the result. Always inspect the results of merge() to make sure it is doing what you expect (i.e., that you wrote the code correctly to get what you wanted!).
The commands below duplicate the merging done piecemeal above.
# x2 contains the response variable
dat <- x2
# merge by the variables in common:
dat2 <- merge(dat, x1)
dat3 <- merge(dat2, x3)
head(dat3)## loc uocc ind year doy lat long basin
## 1 RV_01_16345 occ_001 48.40576 2017 54 34.05400 -83.24100 Savannah (01)
## 2 RV_01_240 occ_002 56.78960 2006 25 32.67461 -81.46906 Savannah (01)
## 3 RV_01_241 occ_003 60.06336 2003 33 33.58468 -82.65235 Savannah (01)
## 4 RV_01_242 occ_004 21.30232 2002 292 33.38387 -82.00422 Savannah (01)
## 5 RV_01_243 occ_005 51.50136 2002 12 33.83757 -82.91251 Savannah (01)
## 6 RV_01_244 occ_006 73.91343 2007 16 34.95895 -83.57158 Savannah (01)
## huc12 state county ecoregion3 ecoregion4
## 1 30601040402 Georgia Madison Piedmont 45b
## 2 30601090104 Georgia Screven Southern Coastal Plain 75f
## 3 30601050203 Georgia Warren Piedmont 45c
## 4 30601060602 Georgia Richmond Southeastern Plains 65c
## 5 30601040504 Georgia Wilkes Piedmont 45c
## 6 30601020101 Georgia Towns Blue Ridge 66d
## eco4 gridcode catchkm2 fid bfi
## 1 Southern Outer Piedmont 2331987 2.3616 6278849 59.00000
## 2 Sea Island Flatwoods 2339456 12.5667 20103925 57.01117
## 3 Carolina Slate Belt 2334311 11.7000 6289897 44.00000
## 4 Sand Hills 2335367 1.8666 22725381 63.94214
## 5 Carolina Slate Belt 2331832 13.6170 6280051 50.75585
## 6 Southern Crystaline Ridges and Mountains 2328238 4.6449 6267588 64.17283
## elev roaddens storder flow slope huc08 fish.rich
## 1 206.97063 2.4394053 3 -0.24495427 0.00001000 3060104 35
## 2 28.11548 2.1069683 3 -0.23087494 0.00176311 3060109 61
## 3 142.94652 1.6804648 1 -0.81467829 0.00877661 3060105 21
## 4 55.97662 3.4675126 3 -0.03230713 0.00055628 3060106 83
## 5 166.52806 1.5691058 1 -0.72841189 0.00605539 3060104 35
## 6 925.20189 0.3386832 1 -0.52734597 0.03813850 3060102 51# what if you only want some variables to be merged?
x3.cols <- c("loc", "lat", "long")
dat4 <- merge(x2, x1)
dat4 <- merge(dat4, x3[,x3.cols])
head(dat4)## loc uocc ind year doy lat long
## 1 RV_01_16345 occ_001 48.40576 2017 54 34.05400 -83.24100
## 2 RV_01_240 occ_002 56.78960 2006 25 32.67461 -81.46906
## 3 RV_01_241 occ_003 60.06336 2003 33 33.58468 -82.65235
## 4 RV_01_242 occ_004 21.30232 2002 292 33.38387 -82.00422
## 5 RV_01_243 occ_005 51.50136 2002 12 33.83757 -82.91251
## 6 RV_01_244 occ_006 73.91343 2007 16 34.95895 -83.57158Some of the tidyverse packages offer alternative solutions for merging datasets that can be more or less elegant than merge(). However, the methods shown above use only base R functions and will get the job done.
3.5.4 Reshaping data frames
In your career you are likely to encounter datasets organized in a variety of formats. Most of these formats fall into one of two categories: wide and long.
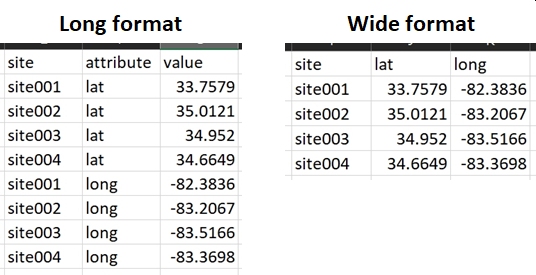
In a long-format dataset, every value is stored in a separate row. The variable (AKA: feature or attribute) that each value represents is kept in a separate column. Many public or online data sources serve datasets in long format. This is usually because the data are pulled from a database, and it left to the user to reshape the data to fit their needs. Storing data in this manner can sometimes be more efficient from a computational standpoint, but it is usually not convenient for analysis. The reason that long format is inconvenient for analysis is because all of the information about individual samples (i.e, observations) are on different rows of the dataset.
In a wide-format dataset, each row contains all values associated with a particular sample. This means that each variable gets its own column, and each cell contains a single value. This format is sometimes called tidy format by the authors of the tidyverse set of R packages (Wickham 2014), but researchers were formatting data this way long before the tidyverse packages or R were developed (Broman and Woo 2018). As a rule, you should store and manage your data in wide/tidy format. This format makes it easy to take advantage of R’s vectorized nature.
Base R has some functions for reshaping data, but most sources recommend using the reshape2 package or the tidyverse. The examples below demonstrate reshaping a dataset from a typical long format to a more useful wide format. You will need the file reshape_example.csv in your R home directory and package reshape2.
## Warning: package 'reshape2' was built under R version 4.3.3## site attribute value
## 1 RV_01_245 lat 33.757913
## 2 RV_01_245 long -82.383578
## 3 RV_01_245 catchkm2 11.079
## 4 RV_01_245 ecoregion3 Piedmont
## 5 RV_01_248 lat 34.952033
## 6 RV_01_248 long -83.516598## [1] "character"Notice that the value column contains a mix of numeric and text values. This means that the numbers are stored as text. If we wanted to do something with those values, they need to be pulled out of the vector value and converted to numeric values. Because a vector can have only one type of value, the attributes will need to end up in their own columns. The fastest way to do this is to use the function dcast() from package reshape2.
dx <- dcast(x, site~attribute)
dx$catchkm2 <- as.numeric(dx$catchkm2)
dx$lat <- as.numeric(dx$lat)
dx$long <- as.numeric(dx$long)
head(dx)## site catchkm2 ecoregion3 lat long
## 1 RV_01_245 11.0790 Piedmont 33.75791 -82.38358
## 2 RV_01_248 7.5924 Blue Ridge 34.95203 -83.51660
## 3 RV_01_250 0.6165 Blue Ridge 34.66491 -83.36978
## 4 RV_01_253 2.5614 Piedmont 33.84107 -82.95028
## 5 RV_01_255 7.8687 Piedmont 33.75356 -82.54828
## 6 RV_01_257 1.9629 Piedmont 33.66435 -82.55340If we needed to go the other way, from wide to long format, we use function melt(). This is essentially the reverse of dcast().
## site variable value
## 1 RV_01_245 lat 33.757913
## 2 RV_01_248 lat 34.952033
## 3 RV_01_250 lat 34.664911
## 4 RV_01_253 lat 33.841068
## 5 RV_01_255 lat 33.753558
## 6 RV_01_257 lat 33.664353## site variable value
## 1 RV_01_245 lat 33.757913
## 11 RV_01_245 long -82.383578
## 21 RV_01_245 catchkm2 11.079
## 31 RV_01_245 ecoregion3 Piedmont
## 2 RV_01_248 lat 34.952033
## 12 RV_01_248 long -83.516598The tidyverse equivalents for melt() and dcast() are pivot_longer() and pivot_wider(), respectively. The syntax of these tidyverse functions are similar to the reshape2 functions. Note that the tidyverse functions will output a tibble, which is a form of data frame peculiar to the tidyverse. Some non-tidyverse functions can use tibbles, and some cannot. Tibbles can be converted to base R data frames with function data.frame().
## Warning: package 'tidyr' was built under R version 4.3.3##
## Attaching package: 'tidyr'## The following object is masked from 'package:reshape2':
##
## smiths## # A tibble: 6 x 5
## site lat long catchkm2 ecoregion3
## <chr> <chr> <chr> <chr> <chr>
## 1 RV_01_245 33.757913 -82.383578 11.079 Piedmont
## 2 RV_01_248 34.952033 -83.516598 7.5924 Blue Ridge
## 3 RV_01_250 34.664911 -83.369779 0.6165 Blue Ridge
## 4 RV_01_253 33.841068 -82.95028 2.5614 Piedmont
## 5 RV_01_255 33.753558 -82.548275 7.8687 Piedmont
## 6 RV_01_257 33.664353 -82.553397 1.9629 Piedmonttmx <- pivot_longer(tdx,
cols=c("lat", "long", "catchkm2", "ecoregion3"),
names_to="variable")
head(tmx)## # A tibble: 6 x 3
## site variable value
## <chr> <chr> <chr>
## 1 RV_01_245 lat 33.757913
## 2 RV_01_245 long -82.383578
## 3 RV_01_245 catchkm2 11.079
## 4 RV_01_245 ecoregion3 Piedmont
## 5 RV_01_248 lat 34.952033
## 6 RV_01_248 long -83.516598Literature Cited
I don’t know if there is an actual name for this.↩︎
A binary operator uses two arguments, one before the symbol and one after. For example,
+is a binary operator that adds the the argument before the+to the argument after the+.↩︎This is slightly longer than a Rent year, which is 525600 minutes (31536000 seconds).↩︎
cbind()also works on matrices.↩︎